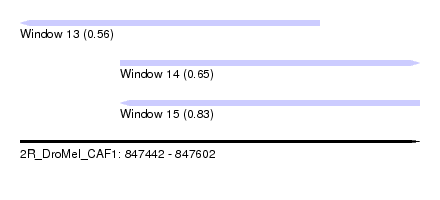
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 847,442 – 847,602 |
| Length | 160 |
| Max. P | 0.833083 |

| Location | 847,442 – 847,562 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.53 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -21.42 |
| Energy contribution | -22.05 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 847442 120 - 20766785 AUUGAAUUAUUUGAUAAGCUUUCUUUGCGCUUUAACGAACGAAUUUUGUUUAUUAAAGACGUAAUUGGGCCAAGUGAAAUCUGCUGCUGGUCAUUUUACGGACACGGAAAAAAAGUAGCU ......(((((((....((((...(((((((((((..(((((...)))))..)))))).)))))..))))))))))).....((((((.....((((.((....)).))))..)))))). ( -29.50) >DroSim_CAF1 307 120 - 1 AUUGAACUAUUUGAUAAGCUUUCGUUGCGCUUUAAUGAACGAAUUUUGUUCAUUAAAGAUGUAAUUGGACCCAGUGAAAUCUGCUGCUGGUCAUUUUACGGACAUGGAAAAAAAGUAGCU .......................(((((.(((((((((((((...))))))))))))).(((((...((((((((.......))))..))))...)))))..............))))). ( -30.10) >DroYak_CAF1 34998 120 - 1 AUUGAACUAUUUGAUAAGCUUUCUUUGCGCUUUAAUGAACGAAUUUUGUUCAUUAAAGACGUAAUUGGACCUAGUGAAAUCUGUUGCUGGUCAUUUUACGGACACGGCAAAAAAGUGGCU .................(((((.(((((.(((((((((((((...))))))))))))).(((((...((((.((..(......)..))))))...)))))......)))))))))).... ( -35.80) >DroMoj_CAF1 2387 120 - 1 AUUGAACUGUUCGACAAGCUAUCGUUGCGCUUCAACGAACGAAUACUUUUUAUUAAGGACGUCAUCGGUCCAAGUGAGAUCUGUUGCUGGUCCUUCUAUGGUCAUGGCAAGAAAGUAGCG .......((((((..((((.........))))...))))))..((((((((.....((((.......)))).............((((((.((......)).).)))))))))))))... ( -26.10) >consensus AUUGAACUAUUUGAUAAGCUUUCGUUGCGCUUUAACGAACGAAUUUUGUUCAUUAAAGACGUAAUUGGACCAAGUGAAAUCUGCUGCUGGUCAUUUUACGGACACGGAAAAAAAGUAGCU ............(((..((((((((((((((((((((((((.....)))))))))))).))))).))))...)))...))).((((((.(((........)))..........)))))). (-21.42 = -22.05 + 0.63)

| Location | 847,482 – 847,602 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.39 |
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -18.36 |
| Energy contribution | -17.80 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 847482 120 + 20766785 AUUUCACUUGGCCCAAUUACGUCUUUAAUAAACAAAAUUCGUUCGUUAAAGCGCAAAGAAAGCUUAUCAAAUAAUUCAAUAUUUUUUAAUAAGUUGUCGUCUGAUGUACUUGUGUACGAA ...(((..((((.......((.((((((..(((.......)))..)))))))).......(((((((.(((.(((.....))).))).)))))))))))..)))(((((....))))).. ( -21.70) >DroSim_CAF1 347 120 + 1 AUUUCACUGGGUCCAAUUACAUCUUUAAUGAACAAAAUUCGUUCAUUAAAGCGCAACGAAAGCUUAUCAAAUAGUUCAAUAUUUUUUAAUAAGUUGUCGUCUGAUGUACUUGUGUACGAA ..........((((((.((((((((((((((((.......)))))))))).....((((.(((((((.(((.(((.....))).))).))))))).))))..)))))).))).).))... ( -27.20) >DroYak_CAF1 35038 120 + 1 AUUUCACUAGGUCCAAUUACGUCUUUAAUGAACAAAAUUCGUUCAUUAAAGCGCAAAGAAAGCUUAUCAAAUAGUUCAAUAUUUUUUAAUAAGUUGUCAUCUGAUGUACUUGUGUAUGAA ....(((.((((.((....((.(((((((((((.......)))))))))))))....((.(((((((.(((.(((.....))).))).))))))).))......)).)))))))...... ( -25.40) >DroMoj_CAF1 2427 120 + 1 AUCUCACUUGGACCGAUGACGUCCUUAAUAAAAAGUAUUCGUUCGUUGAAGCGCAACGAUAGCUUGUCGAACAGUUCAAUAUUCUUGAGCAAAUUAUCAUCCGAUGUGCUUGUGUACGAG ..(((..(((((..(((((.....................(((((...((((.........))))..))))).((((((.....))))))...))))).))))).((((....))))))) ( -28.30) >consensus AUUUCACUUGGUCCAAUUACGUCUUUAAUAAACAAAAUUCGUUCAUUAAAGCGCAAAGAAAGCUUAUCAAAUAGUUCAAUAUUUUUUAAUAAGUUGUCAUCUGAUGUACUUGUGUACGAA ..(((........((....((.(((((((((((.......)))))))))))))....((.(((((((.(((.(((.....))).))).))))))).))...))..((((....))))))) (-18.36 = -17.80 + -0.56)

| Location | 847,482 – 847,602 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.39 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -20.60 |
| Energy contribution | -21.35 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 847482 120 - 20766785 UUCGUACACAAGUACAUCAGACGACAACUUAUUAAAAAAUAUUGAAUUAUUUGAUAAGCUUUCUUUGCGCUUUAACGAACGAAUUUUGUUUAUUAAAGACGUAAUUGGGCCAAGUGAAAU ...((((....)))).(((.........((((((((.(((.....))).))))))))((((...(((((((((((..(((((...)))))..)))))).)))))..))))....)))... ( -24.70) >DroSim_CAF1 347 120 - 1 UUCGUACACAAGUACAUCAGACGACAACUUAUUAAAAAAUAUUGAACUAUUUGAUAAGCUUUCGUUGCGCUUUAAUGAACGAAUUUUGUUCAUUAAAGAUGUAAUUGGACCCAGUGAAAU ...((((....))))....(((((...(((((((((.............)))))))))...)))))((((((((((((((((...))))))))))))).))).((((....))))..... ( -28.92) >DroYak_CAF1 35038 120 - 1 UUCAUACACAAGUACAUCAGAUGACAACUUAUUAAAAAAUAUUGAACUAUUUGAUAAGCUUUCUUUGCGCUUUAAUGAACGAAUUUUGUUCAUUAAAGACGUAAUUGGACCUAGUGAAAU ......(((((((..((((((((......((((....))))......))))))))..))))(((((((((((((((((((((...))))))))))))).)))))..)))....))).... ( -28.80) >DroMoj_CAF1 2427 120 - 1 CUCGUACACAAGCACAUCGGAUGAUAAUUUGCUCAAGAAUAUUGAACUGUUCGACAAGCUAUCGUUGCGCUUCAACGAACGAAUACUUUUUAUUAAGGACGUCAUCGGUCCAAGUGAGAU .((((.....(((...((((((((((.(((.....))).)))).....))))))...))).((((((.....)))))))))).....(((((((..((((.......)))).))))))). ( -22.70) >consensus UUCGUACACAAGUACAUCAGACGACAACUUAUUAAAAAAUAUUGAACUAUUUGAUAAGCUUUCGUUGCGCUUUAACGAACGAAUUUUGUUCAUUAAAGACGUAAUUGGACCAAGUGAAAU ......(((.(((..((((((((......((((....))))......))))))))..)))(((((((((((((((((((((.....)))))))))))).))))).))))....))).... (-20.60 = -21.35 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:23:13 2006