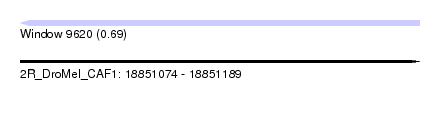
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,851,074 – 18,851,189 |
| Length | 115 |
| Max. P | 0.689774 |

| Location | 18,851,074 – 18,851,189 |
|---|---|
| Length | 115 |
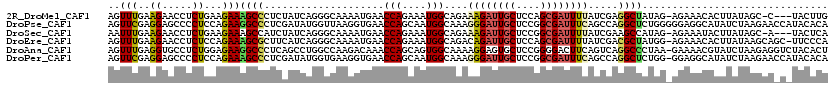
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.87 |
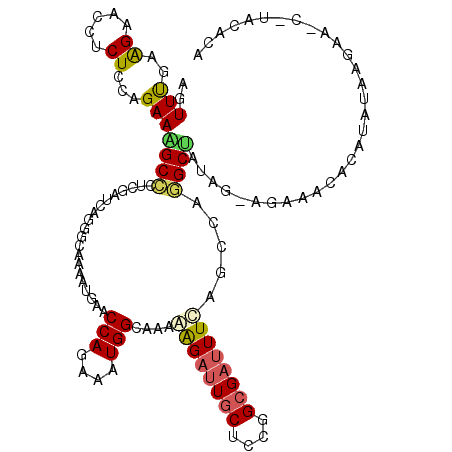
| Mean single sequence MFE | -36.07 |
| Consensus MFE | -12.52 |
| Energy contribution | -12.25 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18851074 115 - 20766785 AGUUUGAAGAACCUCUGAAGAAAGCCCUCUAUCAGGGCAAAAUGAACCAGAAAUGGCAGAAAGAUUGCUCCAGCGAUUUUAUCGAGGCUAUAG-AGAAACACUUAUAGC-C---UACUUG ...((((......((((......(((((.....)))))........(((....)))))))((((((((....)))))))).))))((((((((-........)))))))-)---...... ( -33.60) >DroPse_CAF1 30460 120 - 1 AGUUCGAGGAGCCCCUCCAGAAGGCCCUCGAUAUGGUUAAGGUGAACCAGCAAUGGCAAAGGGAUUGCUCCGGCGAUUUCAGCCAGGCUCUGGGGGAGGCAUAUCUAAGAACCAUACACA .((((..((((((((((((((..(((...(...(((((......))))).)..((((...(..(((((....)))))..).))))))))))))))).)))...)))..))))........ ( -47.20) >DroSec_CAF1 25243 115 - 1 AAUUUGAAGAACCUCUGAAGAAAGCCAUCUAUCAGGGCAAAAUGAACCAGAAAUGGCAGAAAGAUUGCUCCGGCGAUUUUAUCGAAGCCAUAG-AGAAAUACUUAUAGC-A---UACUCA ......(((...(((((((((......))).))))))...............(((((.((((((((((....)))))))).))...)))))..-.......))).....-.---...... ( -22.50) >DroEre_CAF1 28870 118 - 1 AGUUUGAAGAACCUCUCCAGAAAGCGCUUCAUCAGGGCAAAAUGAACCAGAAAUGGCAGACAGAUUGCUCCAGCGAUUUUAUCGACGCUAUGG-AGAAACACUUAUAAGCAGC-UUCCCA .((((((((....((((((...((((........((((((..((..(((....)))....))..)))).))..((((...)))).)))).)))-)))....))).)))))...-...... ( -30.50) >DroAna_CAF1 23794 119 - 1 AGUUUGAGGUGCCUCUGGAGAAGGCCCUCAGCCUGGCCAAGACAAACCAGCAGUGGCAAAAGGAGUGCUCCGGGGACUUCAGUCAGGCCCUAA-GAAAACGUAUCUAAGAGGUCUACACU ....((((((.((.((((((.((((.....)))).((((...(......)...))))..........))))))))))))))((.(((((((.(-((.......))).)).)))))))... ( -38.80) >DroPer_CAF1 30570 119 - 1 AGUUCGAGGAGCCCCUCCAGAAAGCCCUCGAUAUGGUGAAGGUGAACCAGCAAUGGCAAAGGGAUUGCUCCGGCGAUUUCAGCCAGGCUCUGG-GGAGGCAUAUCUAAGAACCAUACACA .((((..((((((.(((((((..(((...(...((((........)))).)..((((...(..(((((....)))))..).))))))))))))-)).)))...)))..))))........ ( -43.80) >consensus AGUUUGAAGAACCUCUCCAGAAAGCCCUCGAUCAGGGCAAAAUGAACCAGAAAUGGCAAAAAGAUUGCUCCGGCGAUUUCAGCCAGGCUAUAG_AGAAACACAUAUAAGAA_C_UACACA ..(((..((.....))...)))((((....................(((....)))....((((((((....)))))))).....))))............................... (-12.52 = -12.25 + -0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:00 2006