
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,846,997 – 18,847,169 |
| Length | 172 |
| Max. P | 0.966972 |

| Location | 18,846,997 – 18,847,106 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 80.86 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -17.78 |
| Energy contribution | -18.00 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18846997 109 - 20766785 GGAAUCACUCGACACCUCAAUAAAUAUUGAGAGUGUCAGCCACUUGAUGCUAAUGCAAAUUCGACUCGCCUAAGUCAUGUAACCCAUGUCCAUGUCCAUGCCCAUGC-CC .((((.....(((((((((((....)))))).)))))..........(((....))).))))((((......))))........((((..(((....)))..)))).-.. ( -26.10) >DroPse_CAF1 26619 98 - 1 GGCACUCCUCGACACCUCUAUAAAUAUUGAGAGUGUCAGGCACUUGAUGCUAAUGCAAAUUCAGUCCGC--------UGCCAGCCAUGUCCGUAAC----CCCAUGCCUC ((((......((((((((..........))).))))).(((......(((....)))....(((....)--------))...)))...........----....)))).. ( -25.80) >DroSim_CAF1 21210 109 - 1 GGAAUCACUCGACACCUCAAUAAAUAUUGAGAGUGUCAGCCACUUGAUGCUAAUGCAAAUUCGACUCGCCUAAGUCAUGUAACCCAUGUCCAUGCCCAUGCCCAUGC-CC .((((.....(((((((((((....)))))).)))))..........(((....))).))))((((......))))........((((..(((....)))..)))).-.. ( -26.10) >DroEre_CAF1 24844 97 - 1 GGAAUCACUCGACACCUCAAUAAAUAUUGAGAGUGUCAGCCACUUGAUGCUAAUGCAAAUUCGACUCGCCUAAGUCAUGUAACCCAUGUCCAU------------GU-CC (((.......(((((((((((....)))))).)))))..........(((....))).....((((......))))............)))..------------..-.. ( -22.80) >DroYak_CAF1 21303 97 - 1 GGAAUCACUCGACACCUCAAUAAAUAUUGAGAGUGUCAGCCACUUGAUGCUAAUGCAAAUUCGACUUGCCUAAGUCAUGUAACCCAUGUCCAU------------GC-CC (((.......(((((((((((....)))))).)))))..........(((....))).....(((((....)))))............)))..------------..-.. ( -23.50) >DroPer_CAF1 26757 98 - 1 GGCACUCCUCGACACCUCUAUAAAUAUUGAGAGUGUCAGGCACUUGAUGCUAAUGCAAAUUCAGUCCGC--------UGCCAGCCAUGUCCGUAAC----CCCAUGCCUC ((((......((((((((..........))).))))).(((......(((....)))....(((....)--------))...)))...........----....)))).. ( -25.80) >consensus GGAAUCACUCGACACCUCAAUAAAUAUUGAGAGUGUCAGCCACUUGAUGCUAAUGCAAAUUCGACUCGCCUAAGUCAUGUAACCCAUGUCCAU__C____CCCAUGC_CC (((.......(((((((((((....)))))).)))))......(((((((....)))...))))........................)))................... (-17.78 = -18.00 + 0.22)

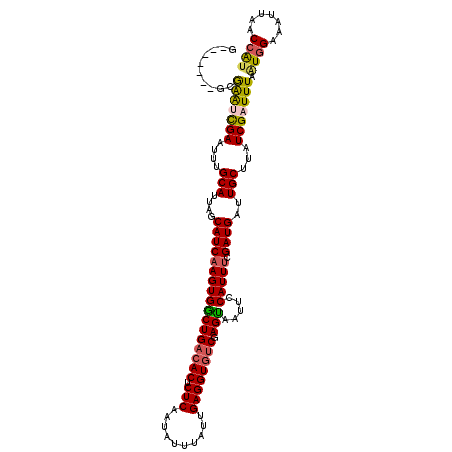
| Location | 18,847,036 – 18,847,146 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 86.82 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -25.68 |
| Energy contribution | -25.18 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
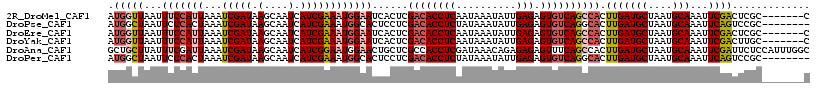
>2R_DroMel_CAF1 18847036 110 + 20766785 G-------GCGAGUCGAAUUUGCAUUAGCAUCAAGUGGCUGACACUCUCAAUAUUUAUUGAGGUGUCGAGUGAUUCCAUUUCGAUGAUUGCUUAUCGAUUUAAUGGAAAUUAACCAU (-------(..(((((....(((....))).....)))))(((((.((((((....)))))))))))......((((((((((((((....)))))))...))))))).....)).. ( -33.90) >DroPse_CAF1 26648 109 + 1 --------GCGGACUGAAUUUGCAUUAGCAUCAAGUGCCUGACACUCUCAAUAUUUAUAGAGGUGUCGAGGAGUGCCAUUUCGAUGAUUGCUUAUCGAUUUAGUGGGAAUUAGCCAU --------..((.((((.(((.(((((((((......((((((((.(((..........)))))))).))).))))....(((((((....)))))))..))))).))))))))).. ( -31.90) >DroEre_CAF1 24871 110 + 1 G-------GCGAGUCGAAUUUGCAUUAGCAUCAAGUGGCUGACACUCUCAAUAUUUAUUGAGGUGUCGAGUGAUUCCAUUUCGAUGAUUGCUUAUCGAUUUAAUGGAAAUUAACCAU (-------(..(((((....(((....))).....)))))(((((.((((((....)))))))))))......((((((((((((((....)))))))...))))))).....)).. ( -33.90) >DroYak_CAF1 21330 110 + 1 G-------GCAAGUCGAAUUUGCAUUAGCAUCAAGUGGCUGACACUCUCAAUAUUUAUUGAGGUGUCGAGUGAUUCCAUUUCGAUGAUUGCUUAUCGAUUUAAUGGAAAUUAACCAU (-------(..(((((....(((....))).....)))))(((((.((((((....)))))))))))......((((((((((((((....)))))))...))))))).....)).. ( -33.80) >DroAna_CAF1 20687 117 + 1 GCCAAAUGGAGAAUCGAAUUUGCAUUAGCAUCAAGUGGCUGAAACUCUCUCUGUUUAUCGAGGUGGCGAGCAGUUCCAUUCCGAUGAUUGCUUAUCGAUUUAAUCGAAAUAAGCAGC ...(((((((((...(......).(((((........))))).....)))))))))((((..((((.........))))..))))..((((((((((((...)))))..))))))). ( -27.50) >DroPer_CAF1 26786 109 + 1 --------GCGGACUGAAUUUGCAUUAGCAUCAAGUGCCUGACACUCUCAAUAUUUAUAGAGGUGUCGAGGAGUGCCAUUUCGAUGAUUGCUUAUCGAUUUAGUGGGAAUUAGCCAU --------..((.((((.(((.(((((((((......((((((((.(((..........)))))))).))).))))....(((((((....)))))))..))))).))))))))).. ( -31.90) >consensus G_______GCGAAUCGAAUUUGCAUUAGCAUCAAGUGGCUGACACUCUCAAUAUUUAUUGAGGUGUCGAGUAAUUCCAUUUCGAUGAUUGCUUAUCGAUUUAAUGGAAAUUAACCAU ..........(((((((....(((....(((((((((((((((((.(((..........)))))))).))).....))))).))))..)))...))))))).((((.......)))) (-25.68 = -25.18 + -0.50)
| Location | 18,847,036 – 18,847,146 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 86.82 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -21.23 |
| Energy contribution | -22.23 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18847036 110 - 20766785 AUGGUUAAUUUCCAUUAAAUCGAUAAGCAAUCAUCGAAAUGGAAUCACUCGACACCUCAAUAAAUAUUGAGAGUGUCAGCCACUUGAUGCUAAUGCAAAUUCGACUCGC-------C .(((((...(((((((...(((((.(....).))))))))))))......(((((((((((....)))))).)))))))))).(((((((....)))...)))).....-------. ( -31.90) >DroPse_CAF1 26648 109 - 1 AUGGCUAAUUCCCACUAAAUCGAUAAGCAAUCAUCGAAAUGGCACUCCUCGACACCUCUAUAAAUAUUGAGAGUGUCAGGCACUUGAUGCUAAUGCAAAUUCAGUCCGC-------- ..((((((((..((.....(((((.(....).)))))..((((((.(((.((((((((..........))).)))))))).....).))))).))..)))).))))...-------- ( -24.30) >DroEre_CAF1 24871 110 - 1 AUGGUUAAUUUCCAUUAAAUCGAUAAGCAAUCAUCGAAAUGGAAUCACUCGACACCUCAAUAAAUAUUGAGAGUGUCAGCCACUUGAUGCUAAUGCAAAUUCGACUCGC-------C .(((((...(((((((...(((((.(....).))))))))))))......(((((((((((....)))))).)))))))))).(((((((....)))...)))).....-------. ( -31.90) >DroYak_CAF1 21330 110 - 1 AUGGUUAAUUUCCAUUAAAUCGAUAAGCAAUCAUCGAAAUGGAAUCACUCGACACCUCAAUAAAUAUUGAGAGUGUCAGCCACUUGAUGCUAAUGCAAAUUCGACUUGC-------C .(((((...(((((((...(((((.(....).))))))))))))......(((((((((((....)))))).)))))))))).(((((((....)))...)))).....-------. ( -31.90) >DroAna_CAF1 20687 117 - 1 GCUGCUUAUUUCGAUUAAAUCGAUAAGCAAUCAUCGGAAUGGAACUGCUCGCCACCUCGAUAAACAGAGAGAGUUUCAGCCACUUGAUGCUAAUGCAAAUUCGAUUCUCCAUUUGGC ((((((((..(((((...))))))))))....((((((.(((....((((.....(((........))).)))).....))).....(((....)))..)))))).........))) ( -25.20) >DroPer_CAF1 26786 109 - 1 AUGGCUAAUUCCCACUAAAUCGAUAAGCAAUCAUCGAAAUGGCACUCCUCGACACCUCUAUAAAUAUUGAGAGUGUCAGGCACUUGAUGCUAAUGCAAAUUCAGUCCGC-------- ..((((((((..((.....(((((.(....).)))))..((((((.(((.((((((((..........))).)))))))).....).))))).))..)))).))))...-------- ( -24.30) >consensus AUGGCUAAUUUCCAUUAAAUCGAUAAGCAAUCAUCGAAAUGGAACCACUCGACACCUCAAUAAAUAUUGAGAGUGUCAGCCACUUGAUGCUAAUGCAAAUUCGACUCGC_______C .(((((...(((((((...(((((.(....).))))))))))))......((((((((..........))).)))))))))).(((((((....)))...))))............. (-21.23 = -22.23 + 1.00)


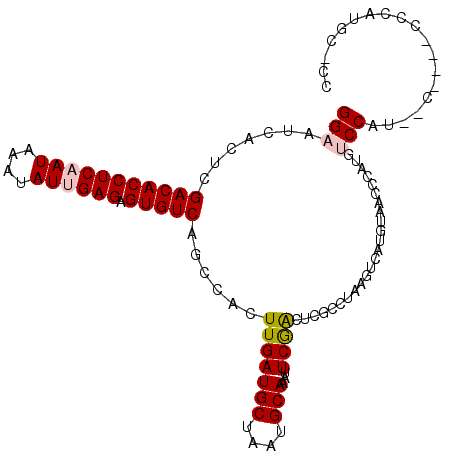
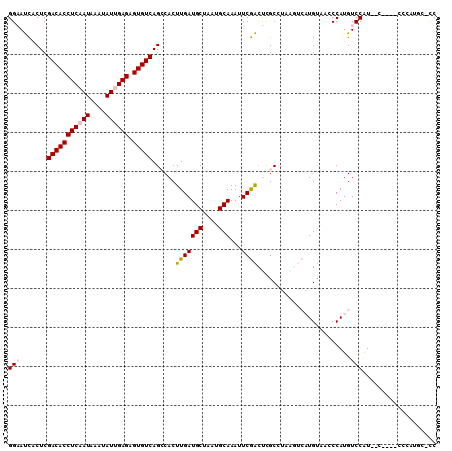
| Location | 18,847,066 – 18,847,169 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.50 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -20.68 |
| Energy contribution | -20.93 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
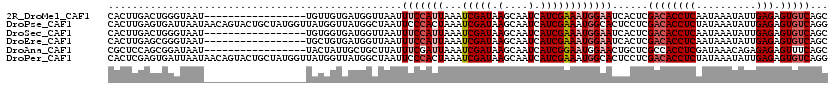
>2R_DroMel_CAF1 18847066 103 + 20766785 GCUGACACUCUCAAUAUUUAUUGAGGUGUCGAGUGAUUCCAUUUCGAUGAUUGCUUAUCGAUUUAAUGGAAAUUAACCAUCACAACA-----------------AUUACCCAGUCAAGUG ((((((((.((((((....)))))))))))..((((((((((((((((((....)))))))...))))))........)))))....-----------------............))). ( -29.20) >DroPse_CAF1 26677 120 + 1 CCUGACACUCUCAAUAUUUAUAGAGGUGUCGAGGAGUGCCAUUUCGAUGAUUGCUUAUCGAUUUAGUGGGAAUUAGCCAUAACCAUAACCAUAGCAGUACUGUUAUUAAUCACUCAAGUG .(((((((.(((..........)))))))).))(((((((((((((((((....)))))))...))))).....................((((((....))))))....)))))..... ( -29.40) >DroSec_CAF1 21015 103 + 1 GCUGACACUCUCAAUAUUUAUUGAGGUGUCGAGUGAUUCCAUUUCGAUGAUUGCUUAUCGAUUUAAUGGAAAUUAACCAUCACCACA-----------------AUUACCCAGUCAAGUG ((((((((.((((((....)))))))))))(.((((((((((((((((((....)))))))...))))))........))))))...-----------------............))). ( -28.80) >DroEre_CAF1 24901 103 + 1 GCUGACACUCUCAAUAUUUAUUGAGGUGUCGAGUGAUUCCAUUUCGAUGAUUGCUUAUCGAUUUAAUGGAAAUUAACCAUCACAGCA-----------------AUUACCCGCUCAAGUG ((((((((.((((((....)))))))))))..((((((((((((((((((....)))))))...))))))........)))))))).-----------------......(((....))) ( -31.90) >DroAna_CAF1 20724 103 + 1 GCUGAAACUCUCUCUGUUUAUCGAGGUGGCGAGCAGUUCCAUUCCGAUGAUUGCUUAUCGAUUUAAUCGAAAUAAGCAGCAAUAGUA-----------------AUUAUCCGCUGGAGCG ((((..((.(((..........)))))..).))).((((((..(.(((((((((((((((((...)))))..))))).((....)))-----------------)))))).).)))))). ( -26.50) >DroPer_CAF1 26815 120 + 1 CCUGACACUCUCAAUAUUUAUAGAGGUGUCGAGGAGUGCCAUUUCGAUGAUUGCUUAUCGAUUUAGUGGGAAUUAGCCAUAACCAUAACCAUAGCAGUACUGUUAUUAAUCACUCGAGUG .(((((((.(((..........)))))))).))(((((((((((((((((....)))))))...))))).....................((((((....))))))....)))))..... ( -29.40) >consensus GCUGACACUCUCAAUAUUUAUUGAGGUGUCGAGUAAUUCCAUUUCGAUGAUUGCUUAUCGAUUUAAUGGAAAUUAACCAUAACAACA_________________AUUACCCACUCAAGUG ((((((((.(((..........)))))))).)))..((((((((((((((....)))))))...)))))))................................................. (-20.68 = -20.93 + 0.25)


| Location | 18,847,066 – 18,847,169 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.50 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -15.72 |
| Energy contribution | -16.75 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18847066 103 - 20766785 CACUUGACUGGGUAAU-----------------UGUUGUGAUGGUUAAUUUCCAUUAAAUCGAUAAGCAAUCAUCGAAAUGGAAUCACUCGACACCUCAAUAAAUAUUGAGAGUGUCAGC .........(((((((-----------------..(....)..)))...(((((((...(((((.(....).))))))))))))..))))(((((((((((....)))))).)))))... ( -27.80) >DroPse_CAF1 26677 120 - 1 CACUUGAGUGAUUAAUAACAGUACUGCUAUGGUUAUGGUUAUGGCUAAUUCCCACUAAAUCGAUAAGCAAUCAUCGAAAUGGCACUCCUCGACACCUCUAUAAAUAUUGAGAGUGUCAGG .....(((((...............(((((((......))))))).........(((..(((((.(....).)))))..))))))))...((((((((..........))).)))))... ( -26.80) >DroSec_CAF1 21015 103 - 1 CACUUGACUGGGUAAU-----------------UGUGGUGAUGGUUAAUUUCCAUUAAAUCGAUAAGCAAUCAUCGAAAUGGAAUCACUCGACACCUCAAUAAAUAUUGAGAGUGUCAGC .........(((((((-----------------..(....)..)))...(((((((...(((((.(....).))))))))))))..))))(((((((((((....)))))).)))))... ( -28.00) >DroEre_CAF1 24901 103 - 1 CACUUGAGCGGGUAAU-----------------UGCUGUGAUGGUUAAUUUCCAUUAAAUCGAUAAGCAAUCAUCGAAAUGGAAUCACUCGACACCUCAAUAAAUAUUGAGAGUGUCAGC ......(((((....)-----------------))))(((((........((((((...(((((.(....).))))))))))))))))..(((((((((((....)))))).)))))... ( -29.70) >DroAna_CAF1 20724 103 - 1 CGCUCCAGCGGAUAAU-----------------UACUAUUGCUGCUUAUUUCGAUUAAAUCGAUAAGCAAUCAUCGGAAUGGAACUGCUCGCCACCUCGAUAAACAGAGAGAGUUUCAGC .(((((((((.(((..-----------------...)))))))(((((..(((((...)))))))))).......))).(((((((.(((..................)))))))))))) ( -22.77) >DroPer_CAF1 26815 120 - 1 CACUCGAGUGAUUAAUAACAGUACUGCUAUGGUUAUGGUUAUGGCUAAUUCCCACUAAAUCGAUAAGCAAUCAUCGAAAUGGCACUCCUCGACACCUCUAUAAAUAUUGAGAGUGUCAGG .....(((((...............(((((((......))))))).........(((..(((((.(....).)))))..))))))))...((((((((..........))).)))))... ( -27.10) >consensus CACUUGAGUGGGUAAU_________________UAUGGUGAUGGCUAAUUUCCAUUAAAUCGAUAAGCAAUCAUCGAAAUGGAACCACUCGACACCUCAAUAAAUAUUGAGAGUGUCAGC .................................................(((((((...(((((.(....).))))))))))))......((((((((..........))).)))))... (-15.72 = -16.75 + 1.03)
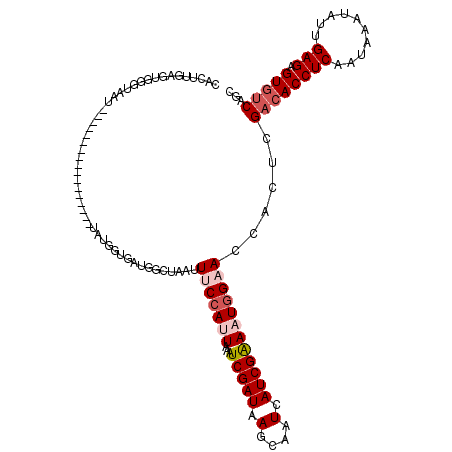
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:59 2006