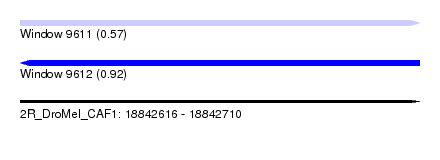
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,842,616 – 18,842,710 |
| Length | 94 |
| Max. P | 0.916702 |

| Location | 18,842,616 – 18,842,710 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 78.18 |
| Mean single sequence MFE | -19.20 |
| Consensus MFE | -11.84 |
| Energy contribution | -11.90 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18842616 94 + 20766785 UGCGAUUCUUUCGCCCUCACUUCG-UC--C-UUUUUUAUGCAUUCGCCUUAAUUGCAUCAACUUGGG-AAUUGUUUGACCAGCGAAUUGACCAAAUAAA .((((.....))))..........-((--(-(..((.(((((...........))))).))...)))-).(((((((..(((....)))..))))))). ( -16.30) >DroSec_CAF1 16645 81 + 1 UGCGAUUCUUUCGC-----------------UUUUUUAUGCAUUUGCCUUAAUUGCAUCAACUUGGG-AAUUGUUUGACCAGCGAAUUGGCCAAAUAAA .((((.....))))-----------------......(((((...........))))).........-..(((((((.((((....)))).))))))). ( -15.80) >DroSim_CAF1 16768 81 + 1 GGCGAUUCUUUCGC-----------------UUUUUUAUGCAUUUGCCUUAAUUGCAUCAACUUGGG-AAUUGUUUGACCAGCGAAUUGGCCAAAUAAA (((.(....(((((-----------------(...(((.(((.((.((................)).-)).))).)))..)))))).).)))....... ( -17.79) >DroEre_CAF1 20528 79 + 1 ----------------UCACUUCG-UC--CUUUUUUUAUGCAUUUCCCUUAAUUGCAUCAACUUGGG-AAUUGUUUGACCAGCGAAUUGGCCAAAUAAA ----------------........-((--((......(((((...........)))))......)))-).(((((((.((((....)))).))))))). ( -16.00) >DroYak_CAF1 17008 97 + 1 UGCGAUUCUUUCGCUCUCACUUCG-UACCAUUUUUUUAUGCAUUUGCCUUAAUUGCAUCAACUUGGG-AAUUGUUUGACCAGCGAAUUGGCCAAAUAAA .((((.....))))..........-..(((.......(((((...........))))).....))).-..(((((((.((((....)))).))))))). ( -18.80) >DroPer_CAF1 22698 97 + 1 UUCUUAUUUUUGGGGGACGGUGCCCCC--CAUUUUUUAUGCAUUUCCCUUAAUUGCAUCAAACUGGGCGAUUGUUUGGCCAGGGAAUUGGCCAAAUAAA ..........((((((.......))))--))..(((.(((((...........))))).)))........((((((((((((....)))))))))))). ( -30.50) >consensus UGCGAUUCUUUCGC__UCACUUCG_UC__C_UUUUUUAUGCAUUUGCCUUAAUUGCAUCAACUUGGG_AAUUGUUUGACCAGCGAAUUGGCCAAAUAAA .((((.....)))).......................(((((...........)))))............(((((((.((((....)))).))))))). (-11.84 = -11.90 + 0.06)

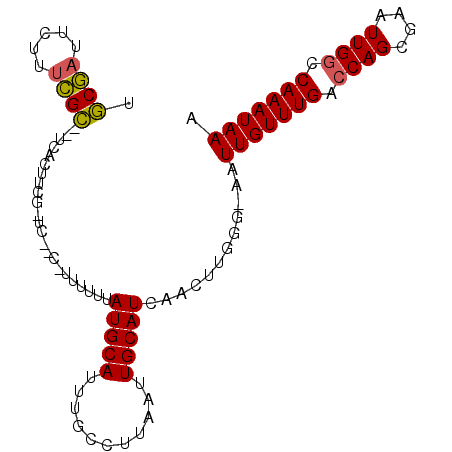
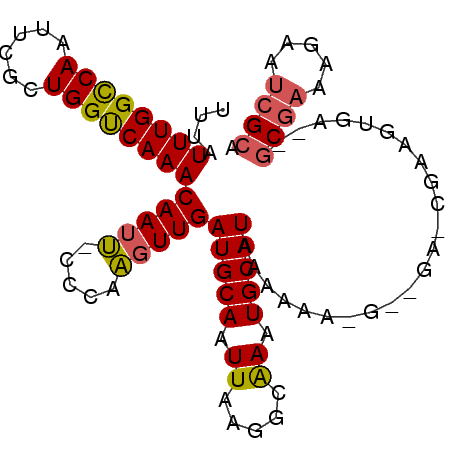
| Location | 18,842,616 – 18,842,710 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 78.18 |
| Mean single sequence MFE | -20.40 |
| Consensus MFE | -13.39 |
| Energy contribution | -13.53 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18842616 94 - 20766785 UUUAUUUGGUCAAUUCGCUGGUCAAACAAUU-CCCAAGUUGAUGCAAUUAAGGCGAAUGCAUAAAAAA-G--GA-CGAAGUGAGGGCGAAAGAAUCGCA (((((((.(((.(((((((.......((((.-.....))))..........)))))))..........-.--))-).))))))).((((.....)))). ( -19.83) >DroSec_CAF1 16645 81 - 1 UUUAUUUGGCCAAUUCGCUGGUCAAACAAUU-CCCAAGUUGAUGCAAUUAAGGCAAAUGCAUAAAAAA-----------------GCGAAAGAAUCGCA ....((((((((......))))))))((((.-.....))))(((((.((.....)).)))))......-----------------((((.....)))). ( -17.90) >DroSim_CAF1 16768 81 - 1 UUUAUUUGGCCAAUUCGCUGGUCAAACAAUU-CCCAAGUUGAUGCAAUUAAGGCAAAUGCAUAAAAAA-----------------GCGAAAGAAUCGCC ....((((((((......))))))))((((.-.....))))(((((.((.....)).)))))......-----------------((((.....)))). ( -17.00) >DroEre_CAF1 20528 79 - 1 UUUAUUUGGCCAAUUCGCUGGUCAAACAAUU-CCCAAGUUGAUGCAAUUAAGGGAAAUGCAUAAAAAAAG--GA-CGAAGUGA---------------- ....((((((((......))))))))((.((-(((.(((((...)))))..))))).))...........--..-........---------------- ( -16.00) >DroYak_CAF1 17008 97 - 1 UUUAUUUGGCCAAUUCGCUGGUCAAACAAUU-CCCAAGUUGAUGCAAUUAAGGCAAAUGCAUAAAAAAAUGGUA-CGAAGUGAGAGCGAAAGAAUCGCA ....((((((((......)))))))).....-.(((..((.(((((.((.....)).)))))...))..)))..-....((((...(....)..)))). ( -22.10) >DroPer_CAF1 22698 97 - 1 UUUAUUUGGCCAAUUCCCUGGCCAAACAAUCGCCCAGUUUGAUGCAAUUAAGGGAAAUGCAUAAAAAAUG--GGGGGCACCGUCCCCCAAAAAUAAGAA ....((((((((......))))))))...((.....((((.(((((.((.....)).)))))...))))(--((((((...)))))))........)). ( -29.60) >consensus UUUAUUUGGCCAAUUCGCUGGUCAAACAAUU_CCCAAGUUGAUGCAAUUAAGGCAAAUGCAUAAAAAA_G__GA_CGAAGUGA__GCGAAAGAAUCGCA ....((((((((......))))))))(((((.....)))))(((((.((.....)).))))).......................((((.....)))). (-13.39 = -13.53 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:53 2006