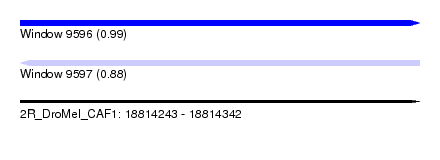
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,814,243 – 18,814,342 |
| Length | 99 |
| Max. P | 0.993172 |

| Location | 18,814,243 – 18,814,342 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 70.75 |
| Mean single sequence MFE | -21.58 |
| Consensus MFE | -10.15 |
| Energy contribution | -11.77 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18814243 99 + 20766785 UUACGAUCUCCGCCGUCUUCGGAUGCACCUCCUCCUCCCCCU-GCUCCUC----CUACGCCGCUC------CGUAGCAAAAUCCAUUUGCAUAUUUGCAUCCUUAUGCCU ..(((........)))....(((((((...............-.......----.((((......------))))(((((.....))))).....)))))))........ ( -17.30) >DroVir_CAF1 13490 104 + 1 CUUCGCCCCCCUCGAGCUUCAAC-ACAAU-ACCCCGCGGGUCCCAUUGCA-UG-CUGC--CACACUGUUCGCUUGGCAAAAUGCAUUUGCAUAUUUGUAUCCUUAUGCCU ........(((.((.........-.....-....)).)))......((((-(.-.(((--((..(.....)..)))))..)))))...(((((..........))))).. ( -20.07) >DroSec_CAF1 6140 90 + 1 UUACGAUCUCCGCCGGCUUCGGAUGCACCUCCUCC----------UCCUC----CUUUGCCGCUC------CGUGGCAAAAUCCAUUUGCAUAUUUGCAUCCUUAUGCCU ..............(((...(((((((........----------.....----.((((((((..------.))))))))...............)))))))....))). ( -23.85) >DroSim_CAF1 6172 93 + 1 UUACGAUCUCCGCCGGCUUCGGAUGCACCUCCUCCUCC-------UCCUC----CUUUGCCGCUC------CGUGGCAAAAUCCAUUUGCAUAUUUGCAUCCUUAUGCCU ..............(((...(((((((...........-------.....----.((((((((..------.))))))))...(....)......)))))))....))). ( -22.30) >DroEre_CAF1 4574 87 + 1 UUACGAUCUCCGCCGGCUUCGGAUGCACCGCCUCC---------------C--ACCCCGCCGCUU------CGUGGCAAAAUCCAUUUGCAUAUUUGCAUCCUUAUGCCU ..............(((...(((((((........---------------.--....(((.....------.)))(((((.....))))).....)))))))....))). ( -20.90) >DroAna_CAF1 728 102 + 1 UUACGA-CUCCUCCAGCUACAGCUACAACUACAGCUCCAGCUUCGUCCCA-GGCCCAGGCCUUGG------CAUGGCAAAAUCCAUUUGCAUAUUUGCAUCCUUAUGCCU ..((((-.......((((..((((........))))..))))))))((.(-(((....)))).))------...((((.........((((....))))......)))). ( -25.07) >consensus UUACGAUCUCCGCCGGCUUCGGAUGCACCUCCUCCUCC_______UCCUC____CUACGCCGCUC______CGUGGCAAAAUCCAUUUGCAUAUUUGCAUCCUUAUGCCU ..............(((...(((((((.................................(((.........)))(((((.....))))).....)))))))....))). (-10.15 = -11.77 + 1.61)

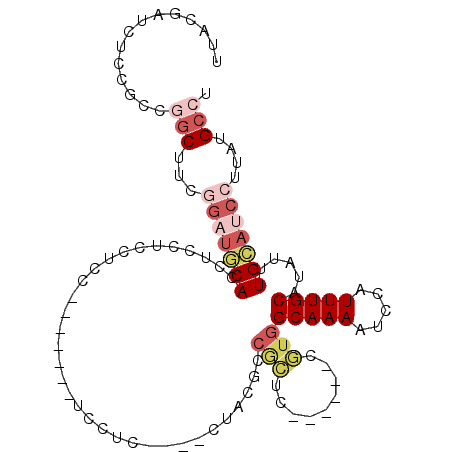
| Location | 18,814,243 – 18,814,342 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 70.75 |
| Mean single sequence MFE | -28.18 |
| Consensus MFE | -12.76 |
| Energy contribution | -12.60 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18814243 99 - 20766785 AGGCAUAAGGAUGCAAAUAUGCAAAUGGAUUUUGCUACG------GAGCGGCGUAG----GAGGAGC-AGGGGGAGGAGGAGGUGCAUCCGAAGACGGCGGAGAUCGUAA ........(((((((..(.(.(...((..((((.(((((------......)))))----.)))).)-)...).).....)..)))))))....((((......)))).. ( -22.60) >DroVir_CAF1 13490 104 - 1 AGGCAUAAGGAUACAAAUAUGCAAAUGCAUUUUGCCAAGCGAACAGUGUG--GCAG-CA-UGCAAUGGGACCCGCGGGGU-AUUGU-GUUGAAGCUCGAGGGGGGCGAAG .........(((((((.........(((((.((((((.((.....)).))--))))-.)-)))).....((((...))))-.))))-)))...((((.....)))).... ( -30.60) >DroSec_CAF1 6140 90 - 1 AGGCAUAAGGAUGCAAAUAUGCAAAUGGAUUUUGCCACG------GAGCGGCAAAG----GAGGA----------GGAGGAGGUGCAUCCGAAGCCGGCGGAGAUCGUAA .(((....(((((((..(...(.......(((((((...------....)))))))----.....----------.)...)..)))))))...))).(((.....))).. ( -26.72) >DroSim_CAF1 6172 93 - 1 AGGCAUAAGGAUGCAAAUAUGCAAAUGGAUUUUGCCACG------GAGCGGCAAAG----GAGGA-------GGAGGAGGAGGUGCAUCCGAAGCCGGCGGAGAUCGUAA .(((....(((((((..(...(.......(((((((...------....)))))))----.....-------....)...)..)))))))...))).(((.....))).. ( -25.99) >DroEre_CAF1 4574 87 - 1 AGGCAUAAGGAUGCAAAUAUGCAAAUGGAUUUUGCCACG------AAGCGGCGGGGU--G---------------GGAGGCGGUGCAUCCGAAGCCGGCGGAGAUCGUAA .(((....(((((((....(((......((((((((...------....))))))))--.---------------....))).)))))))...))).(((.....))).. ( -28.70) >DroAna_CAF1 728 102 - 1 AGGCAUAAGGAUGCAAAUAUGCAAAUGGAUUUUGCCAUG------CCAAGGCCUGGGCC-UGGGACGAAGCUGGAGCUGUAGUUGUAGCUGUAGCUGGAGGAG-UCGUAA ..(((((..........))))).....(((((..((.((------((.((((....)))-).)).)).(((((.(((((......))))).)))))))..)))-)).... ( -34.50) >consensus AGGCAUAAGGAUGCAAAUAUGCAAAUGGAUUUUGCCACG______GAGCGGCGAAG____GAGGA_______GGAGGAGGAGGUGCAUCCGAAGCCGGCGGAGAUCGUAA .(((....(((((((.....(((((.....)))))..((.........)).................................)))))))...))).............. (-12.76 = -12.60 + -0.16)
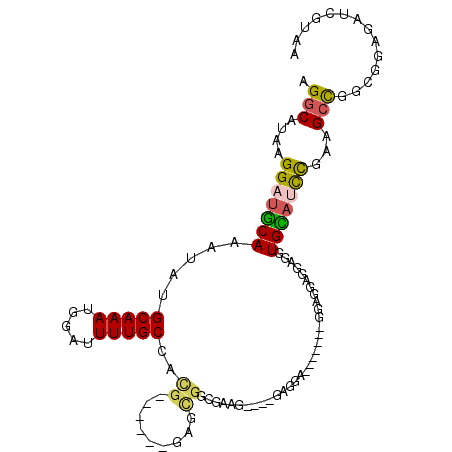
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:38 2006