
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,812,854 – 18,812,956 |
| Length | 102 |
| Max. P | 0.626153 |

| Location | 18,812,854 – 18,812,956 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.64 |
| Mean single sequence MFE | -27.39 |
| Consensus MFE | -10.51 |
| Energy contribution | -11.55 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
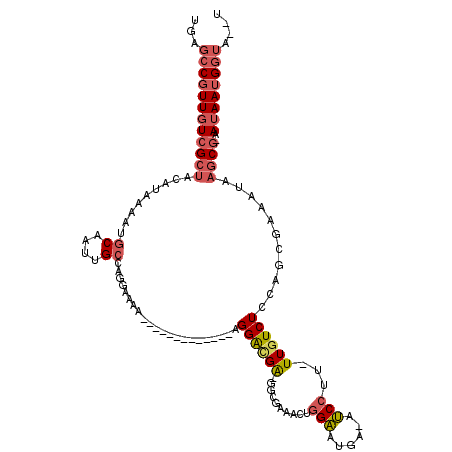
>2R_DroMel_CAF1 18812854 102 + 20766785 UGAGCCGUUGUCGCUACAUAAAAUGCAAUUGCCAGGAAAA-------------AGGACGA-GGCGAAACUGGAAUGA-AUCCUUUUUGUCUUCAACGAAAUAAGCG-AAUAAUGGUA--U ...((((((((((((.........((....))........-------------.(...((-(((((((..(((....-.))).)))))))))...)......))))-.)))))))).--. ( -25.70) >DroVir_CAF1 11851 87 + 1 UGAGCCCUUUUGGGCGUAUGA--UG----------GAGUA-------------AGGGAGGCGGG-----UAGGAUGUCGCCGCU-UUGUCUCGAACGGAAUAAGCGGGAUAAUGAAA--U ...(((((((((..(......--..----------)..))-------------)))).)))...-----.....((((.(((((-(..(((.....)))..))))))))))......--. ( -22.70) >DroSec_CAF1 4668 101 + 1 UGAGCCGUUGUCGCUAAAUAAAAUGCAAUUGCCAGGAAAA-------------AGGAUGA-GGCGAAACGGGAAUGA-AUCCUU-UUGUCUCCAGCGAAAUAAGCG-AAUAAUGGUA--U ...((((((((((((........(((.....((.......-------------.))..((-((((((..((((....-.)))))-)))))))..))).....))))-.)))))))).--. ( -27.42) >DroSim_CAF1 4756 101 + 1 UGAGCCGUUGUCGCUACAUAAAAUGCAAUUGCCAGGAAAA-------------AGGACGA-GGCGAAACUGGAAUGA-AUCCUU-UUGUCUCCAGCGAAAUAAGCG-AAUAAUGGUA--U ...((((((((((((........(((.....((.......-------------.))..((-(((((((..(((....-.)))))-)))))))..))).....))))-.)))))))).--. ( -27.02) >DroEre_CAF1 3248 102 + 1 UGAGCCGUUGUCGCUACAUAAAAUGCAAUUGCCGGGAAAAA------------AGGACGA-GGGAAAACUGGAAUGA-AUCCUU-UUGUCUCCGGCGAAAUAAGCG-AAUAAUGGUA--U ...((((((((((((.(((...)))...(((((((((.(((------------(((((.(-(......)).).....-.)))))-)).)).)))))))....))))-.)))))))).--. ( -34.30) >DroYak_CAF1 3581 116 + 1 UGAGCCGUUGUCGCUACAUAAAAUGCAAUUGCCGGCAAAAAAAAAAAAGAGGAAGGACGA-AGGAAAAGUGGAAUGA-AUCCUU-UUGUCUUCAGCGAAAUAAGCG-AAUAAUGGUAUAU ...((((((((((((........(((........)))...........(..(((((..((-((((............-.)))))-)..)))))..)......))))-.)))))))).... ( -27.22) >consensus UGAGCCGUUGUCGCUACAUAAAAUGCAAUUGCCAGGAAAA_____________AGGACGA_GGCGAAACUGGAAUGA_AUCCUU_UUGUCUCCAGCGAAAUAAGCG_AAUAAUGGUA__U ...((((((((((((.........((....))......................((((((..........(((......)))...))))))...........))))..)))))))).... (-10.51 = -11.55 + 1.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:35 2006