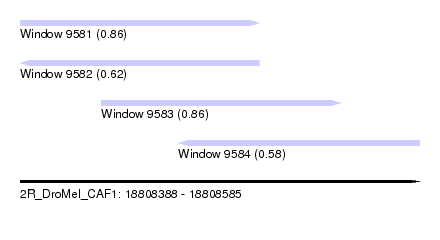
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,808,388 – 18,808,585 |
| Length | 197 |
| Max. P | 0.862482 |

| Location | 18,808,388 – 18,808,506 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 97.74 |
| Mean single sequence MFE | -42.80 |
| Consensus MFE | -41.80 |
| Energy contribution | -42.47 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
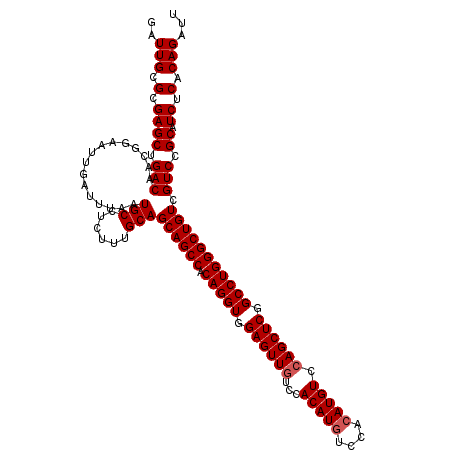
>2R_DroMel_CAF1 18808388 118 + 20766785 GAUUGCGCGAGCUGACAACGGAAUUGAUUUAAUGCCUCUUUGCAGCAGCCACAGGUGGAGUUGUCCACAUGUCCGGAUGUCCAGCUCGGCCUGGGCUGUCGUCCGCAUCUCACAGAUU ..(((.(.((((.(((................(((......)))((((((.(((((.(((((((((........)))....)))))).))))))))))).))).)).)).).)))... ( -41.90) >DroSec_CAF1 301 118 + 1 AAUUGCGCGAGCUGACAACGGAAUUGAUUUAAUGCCUCUUUGCAGCAGCCACAGGUGGAGUUAUCCACAUGUCCACAUGUCCAGCUCGGCCUGGGCUGUCGUCCGCAUCUCACAGAUU ..(((.(.((((.(((................(((......)))((((((.(((((.(((((....(((((....)))))..))))).))))))))))).))).)).)).).)))... ( -41.70) >DroSim_CAF1 361 118 + 1 GAUUGCGCGAGCUGACAACGGAAUUGAUUUAAUGCCUCUUUGCAGCAGCCACAGGUGGAGUUGUCCACAUGUCCACAUGUCCAGCUCGGCCUGGGCUGUCGUCCGCAUCUCACAGAUU ..(((.(.((((.(((................(((......)))((((((.(((((.((((((...(((((....))))).)))))).))))))))))).))).)).)).).)))... ( -44.80) >consensus GAUUGCGCGAGCUGACAACGGAAUUGAUUUAAUGCCUCUUUGCAGCAGCCACAGGUGGAGUUGUCCACAUGUCCACAUGUCCAGCUCGGCCUGGGCUGUCGUCCGCAUCUCACAGAUU ..(((.(.((((.(((................(((......)))((((((.(((((.((((((...(((((....))))).)))))).))))))))))).))).)).)).).)))... (-41.80 = -42.47 + 0.67)

| Location | 18,808,388 – 18,808,506 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 97.74 |
| Mean single sequence MFE | -40.97 |
| Consensus MFE | -40.42 |
| Energy contribution | -40.87 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.619274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18808388 118 - 20766785 AAUCUGUGAGAUGCGGACGACAGCCCAGGCCGAGCUGGACAUCCGGACAUGUGGACAACUCCACCUGUGGCUGCUGCAAAGAGGCAUUAAAUCAAUUCCGUUGUCAGCUCGCGCAAUC ....((((.((.((.((((((.((...((((...((((....))))(((.(((((....))))).)))))))...)).....((.(((.....))).)))))))).)))).))))... ( -40.50) >DroSec_CAF1 301 118 - 1 AAUCUGUGAGAUGCGGACGACAGCCCAGGCCGAGCUGGACAUGUGGACAUGUGGAUAACUCCACCUGUGGCUGCUGCAAAGAGGCAUUAAAUCAAUUCCGUUGUCAGCUCGCGCAAUU ....((((.((.((.((((((((((((((..(((.((..(((((....)))))..)).)))..)))).))))(((.......)))..............)))))).)))).))))... ( -39.90) >DroSim_CAF1 361 118 - 1 AAUCUGUGAGAUGCGGACGACAGCCCAGGCCGAGCUGGACAUGUGGACAUGUGGACAACUCCACCUGUGGCUGCUGCAAAGAGGCAUUAAAUCAAUUCCGUUGUCAGCUCGCGCAAUC ....((((.((.((.((((((((((((((..(((.((..(((((....)))))..)).)))..)))).))))(((.......)))..............)))))).)))).))))... ( -42.50) >consensus AAUCUGUGAGAUGCGGACGACAGCCCAGGCCGAGCUGGACAUGUGGACAUGUGGACAACUCCACCUGUGGCUGCUGCAAAGAGGCAUUAAAUCAAUUCCGUUGUCAGCUCGCGCAAUC ....((((.((.((.((((((((((((((..(((.((..(((((....)))))..)).)))..)))).))))(((.......)))..............)))))).)))).))))... (-40.42 = -40.87 + 0.45)

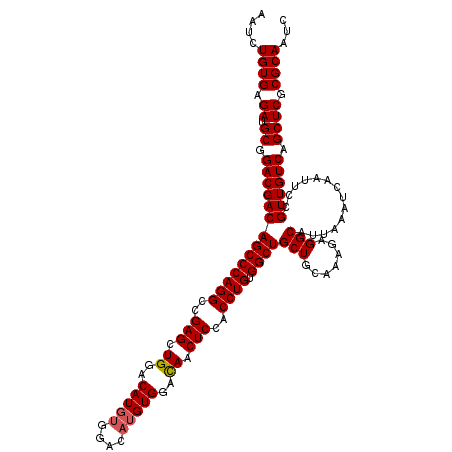
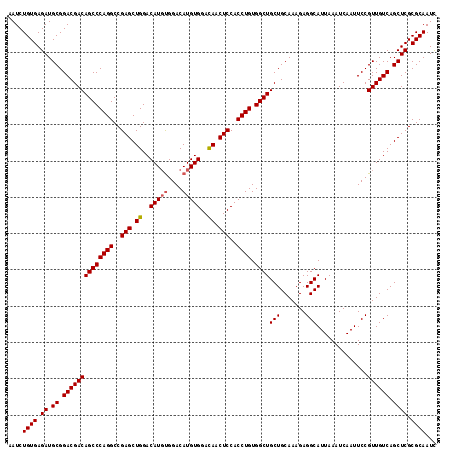
| Location | 18,808,428 – 18,808,546 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 98.31 |
| Mean single sequence MFE | -47.40 |
| Consensus MFE | -46.40 |
| Energy contribution | -47.07 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18808428 118 + 20766785 UGCAGCAGCCACAGGUGGAGUUGUCCACAUGUCCGGAUGUCCAGCUCGGCCUGGGCUGUCGUCCGCAUCUCACAGAUUCGACUGGCUCCUCUUAAUUGGGCCCACUUUGGCGGUAGCG .((.((((((.(((((.(((((((((........)))....)))))).)))))))))))...(((((((.....)))......(((.((........))))).......))))..)). ( -46.50) >DroSec_CAF1 341 118 + 1 UGCAGCAGCCACAGGUGGAGUUAUCCACAUGUCCACAUGUCCAGCUCGGCCUGGGCUGUCGUCCGCAUCUCACAGAUUCGACUGGCUCCUCUUAAUUGGGCCCACUUUGGCGGUAGCG .((.((((((.(((((.(((((....(((((....)))))..))))).)))))))))))...(((((((.....)))......(((.((........))))).......))))..)). ( -46.30) >DroSim_CAF1 401 118 + 1 UGCAGCAGCCACAGGUGGAGUUGUCCACAUGUCCACAUGUCCAGCUCGGCCUGGGCUGUCGUCCGCAUCUCACAGAUUCGACUGGCUCCUCUUAAUUGGGCCCACUUUGGCGGUAGCG .((.((((((.(((((.((((((...(((((....))))).)))))).)))))))))))...(((((((.....)))......(((.((........))))).......))))..)). ( -49.40) >consensus UGCAGCAGCCACAGGUGGAGUUGUCCACAUGUCCACAUGUCCAGCUCGGCCUGGGCUGUCGUCCGCAUCUCACAGAUUCGACUGGCUCCUCUUAAUUGGGCCCACUUUGGCGGUAGCG .((.((((((.(((((.((((((...(((((....))))).)))))).)))))))))))...(((((((.....)))......(((.((........))))).......))))..)). (-46.40 = -47.07 + 0.67)

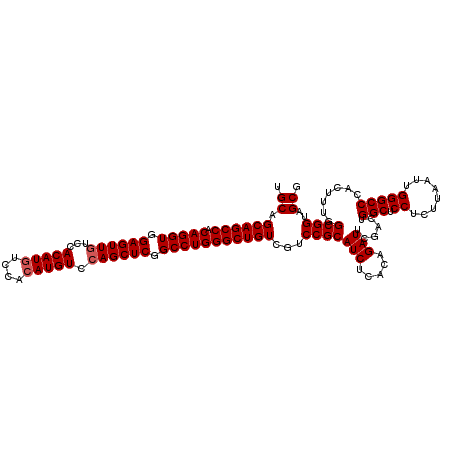
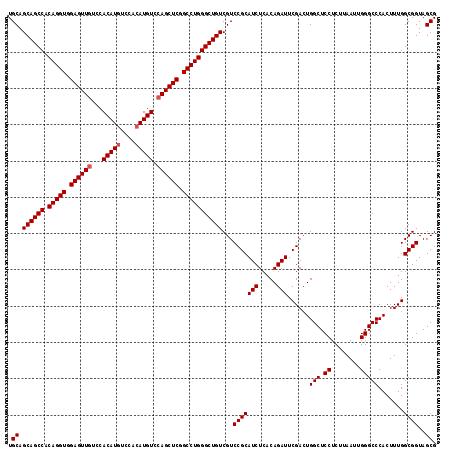
| Location | 18,808,466 – 18,808,585 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -44.30 |
| Consensus MFE | -44.30 |
| Energy contribution | -44.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.38 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18808466 119 - 20766785 GGAUCCGCAAAAAGUGGCCGAGCCACAGGCCUGAAUGAGCGCUACCGCCAAAGUGGGCCCAAUUAAGAGGAGCCAGUCGAAUCUGUGAGAUGCGGACGACAGCCCAGGCCGAGCUGGAC ...(((((.....(((((...))))).((((((..((.(((....))))).....(((((........)).))).((((..(((((.....)))))))))....))))))..)).))). ( -44.30) >DroSec_CAF1 379 119 - 1 GGAUCCGCAAAAAGUGGCCGAGCCACAGGCCUGAAUGAGCGCUACCGCCAAAGUGGGCCCAAUUAAGAGGAGCCAGUCGAAUCUGUGAGAUGCGGACGACAGCCCAGGCCGAGCUGGAC ...(((((.....(((((...))))).((((((..((.(((....))))).....(((((........)).))).((((..(((((.....)))))))))....))))))..)).))). ( -44.30) >DroSim_CAF1 439 119 - 1 GGAUCCGCAAAAAGUGGCCGAGCCACAGGCCUGAAUGAGCGCUACCGCCAAAGUGGGCCCAAUUAAGAGGAGCCAGUCGAAUCUGUGAGAUGCGGACGACAGCCCAGGCCGAGCUGGAC ...(((((.....(((((...))))).((((((..((.(((....))))).....(((((........)).))).((((..(((((.....)))))))))....))))))..)).))). ( -44.30) >consensus GGAUCCGCAAAAAGUGGCCGAGCCACAGGCCUGAAUGAGCGCUACCGCCAAAGUGGGCCCAAUUAAGAGGAGCCAGUCGAAUCUGUGAGAUGCGGACGACAGCCCAGGCCGAGCUGGAC ...(((((.....(((((...))))).((((((..((.(((....))))).....(((((........)).))).((((..(((((.....)))))))))....))))))..)).))). (-44.30 = -44.30 + 0.00)

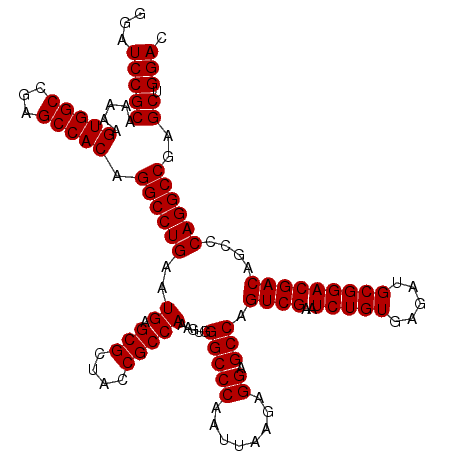
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:26 2006