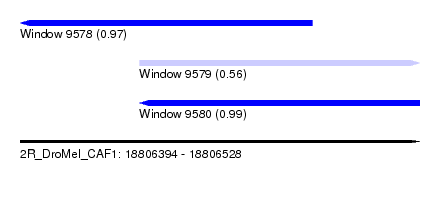
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,806,394 – 18,806,528 |
| Length | 134 |
| Max. P | 0.991332 |

| Location | 18,806,394 – 18,806,492 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 82.60 |
| Mean single sequence MFE | -33.84 |
| Consensus MFE | -20.96 |
| Energy contribution | -21.12 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18806394 98 - 20766785 CGAGGUCCUGG------UUCGGGUCCGGGUCCACGUCCUGAUUCCGGCAUAACAGCCGGAUACGGACCCUUCUGGCCUCUCGCACAAAAUCUACAUUACCAUGA .((((((..((------...(((((((((.......))....((((((......))))))..))))))).)).))))))......................... ( -36.20) >DroSec_CAF1 25687 103 - 1 CGAGGUCCGGGCUCGGGUUCGAGUCCGGUUCCACGUCCUGAUUCCGGCAUAACAGCCGGAUACGGACCCUUCUGGCCUCUCGCACAA-AUCUACAUUACCAUGA .((((((((((((((....)))))))))......((((....((((((......))))))...)))).......)))))........-................ ( -40.60) >DroSim_CAF1 26159 91 - 1 CGGGGUC------------CGGGUCCGGGUCCACGUCCUGAUUCCGGCAUAACAGCCGGAUACGGACCCUUCUGGCCUCUCGCACAA-AUCUACAUUACCAUGA .((((((------------.(((((((((.......))....((((((......))))))..)))))))....))))))........-................ ( -34.50) >DroEre_CAF1 25858 79 - 1 CGGGGU------------------------CCGGGUAAUGAUUCCGGCAUAACAGCCGGAUACGGACCCUUCUGGCCUCUCGCACAA-AUCUACAUUACCAUGA .(((((------------------------(.((((......((((((......)))))).....))))....))))))........-................ ( -26.10) >DroYak_CAF1 26196 80 - 1 CGGGGU------------------------CCGGGUCCUGAUUCCGGCAUAACAGCCGGAUACGGACCCUUCUGGCCUCUCGCACAAAAUCUACAUUACCAUGA .(((((------------------------(.((((((....((((((......))))))...))))))....))))))......................... ( -31.80) >consensus CGGGGUC____________CG_GUCCGG_UCCACGUCCUGAUUCCGGCAUAACAGCCGGAUACGGACCCUUCUGGCCUCUCGCACAA_AUCUACAUUACCAUGA .((((.............................((((....((((((......))))))...))))((....))))))......................... (-20.96 = -21.12 + 0.16)

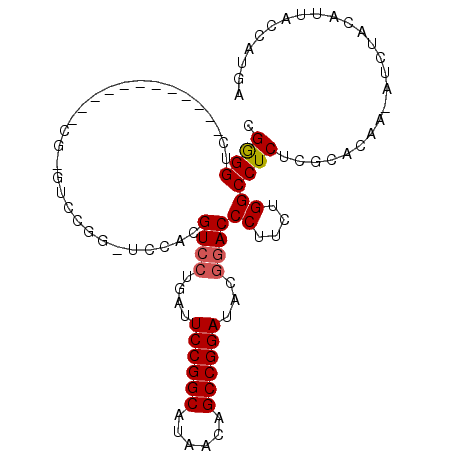
| Location | 18,806,434 – 18,806,528 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 73.34 |
| Mean single sequence MFE | -25.69 |
| Consensus MFE | -12.00 |
| Energy contribution | -12.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18806434 94 + 20766785 CGUAUCCGGCUGUUAUGCCGGAAUCAGGACGUGGACCCGGACCCGAA------CCAGGACCUCGUACCUCGUACCCCGGACUCAAUUACCUCUUGUCUUU ....((((((......))))))((.((((((.((.((.((.......------)).)).)).))).))).)).....((((.............)))).. ( -26.82) >DroSec_CAF1 25726 100 + 1 CGUAUCCGGCUGUUAUGCCGGAAUCAGGACGUGGAACCGGACUCGAACCCGAGCCCGGACCUCGUACCAUGAACCCCUGACUCAAUUACCUCUUGUCUUU ....((((((......)))))).((((((((.((..((((.((((....)))).)))).)).))).)).)))......(((.............)))... ( -35.52) >DroSim_CAF1 26198 74 + 1 CGUAUCCGGCUGUUAUGCCGGAAUCAGGACGUGGACCCGGACCCG------------GACCCCG--------------UACUCAAUUACCUCUUGUCUUU .(((((((((......))))))...((.(((.((..(((....))------------)..))))--------------).))....)))........... ( -22.70) >DroEre_CAF1 25897 76 + 1 CGUAUCCGGCUGUUAUGCCGGAAUCAUUACCCGG------------------------ACCCCGAACCCCGGAGCCCGGACUCAAUUACCUCUUGUCUUU .(((((((((......))))))........((((------------------------.(.(((.....))).).)))).......)))........... ( -23.60) >DroYak_CAF1 26236 76 + 1 CGUAUCCGGCUGUUAUGCCGGAAUCAGGACCCGG------------------------ACCCCGAACCCCGGACCCCGCACUCAAUUACCUCUUGUCUUU .((.((((((......))))))....((..((((------------------------..........))))..)).))..................... ( -19.80) >consensus CGUAUCCGGCUGUUAUGCCGGAAUCAGGACGUGGA_CCGGAC_CG____________GACCCCGAACCCCGGACCCCGGACUCAAUUACCUCUUGUCUUU ....((((((......)))))).............................................................................. (-12.00 = -12.00 + 0.00)



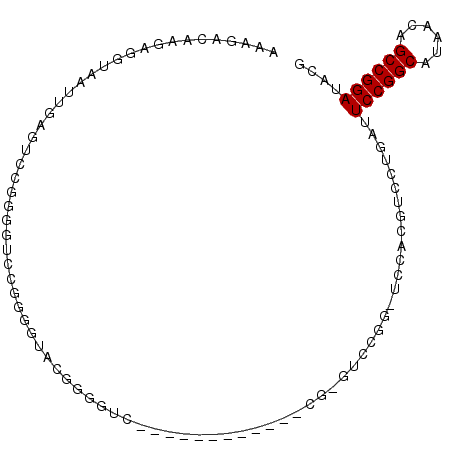
| Location | 18,806,434 – 18,806,528 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 73.34 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -11.20 |
| Energy contribution | -11.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.34 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18806434 94 - 20766785 AAAGACAAGAGGUAAUUGAGUCCGGGGUACGAGGUACGAGGUCCUGG------UUCGGGUCCGGGUCCACGUCCUGAUUCCGGCAUAACAGCCGGAUACG .................(.(((((((((...(((.(((.((.(((((------.......))))).)).)))))).)))))(((......))))))).). ( -33.50) >DroSec_CAF1 25726 100 - 1 AAAGACAAGAGGUAAUUGAGUCAGGGGUUCAUGGUACGAGGUCCGGGCUCGGGUUCGAGUCCGGUUCCACGUCCUGAUUCCGGCAUAACAGCCGGAUACG ...(((..((.....))..)))......(((.((.(((.((.(((((((((....)))))))))..)).)))))))).((((((......)))))).... ( -40.30) >DroSim_CAF1 26198 74 - 1 AAAGACAAGAGGUAAUUGAGUA--------------CGGGGUC------------CGGGUCCGGGUCCACGUCCUGAUUCCGGCAUAACAGCCGGAUACG ...........(((..(.((.(--------------((((..(------------((....)))..)).))).)).).((((((......))))))))). ( -28.60) >DroEre_CAF1 25897 76 - 1 AAAGACAAGAGGUAAUUGAGUCCGGGCUCCGGGGUUCGGGGU------------------------CCGGGUAAUGAUUCCGGCAUAACAGCCGGAUACG ...........(((......(((((((((((.....))))))------------------------))))).......((((((......))))))))). ( -32.10) >DroYak_CAF1 26236 76 - 1 AAAGACAAGAGGUAAUUGAGUGCGGGGUCCGGGGUUCGGGGU------------------------CCGGGUCCUGAUUCCGGCAUAACAGCCGGAUACG ...........(((........((((..(((((.(....).)------------------------))))..))))..((((((......))))))))). ( -30.50) >consensus AAAGACAAGAGGUAAUUGAGUCCGGGGUCCGGGGUACGGGGUC____________CG_GUCCGG_UCCACGUCCUGAUUCCGGCAUAACAGCCGGAUACG ..............................................................................((((((......)))))).... (-11.20 = -11.20 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:22 2006