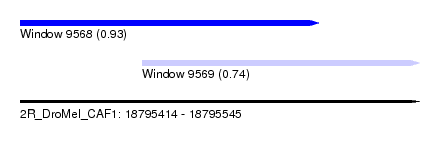
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,795,414 – 18,795,545 |
| Length | 131 |
| Max. P | 0.929712 |

| Location | 18,795,414 – 18,795,512 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.86 |
| Mean single sequence MFE | -22.12 |
| Consensus MFE | -15.11 |
| Energy contribution | -15.67 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18795414 98 + 20766785 CGGUCGCUAAUUGCCUUUGCCAGAAAGUUAAUUACGGCGAA-----------AUUUGAAAAU--UU-CAAAAUUGGACUUGUCAAAAACAUU-UUGACGUUUCCCCUCAGACG------- ..((((.((((((.((((.....)))).))))))))))(((-----------(((....)))--))-)......(((..(((((((.....)-))))))..))).........------- ( -22.20) >DroPse_CAF1 62679 117 + 1 CGGUCGCUAAUUGGCUUUCCCAGA-AGUUAAUUACGAUGAAAAUGUUGAAAACGUUGAAAAU-GCU-GAAAAUUUGACUUGUCAAAAACAUUUUUGACAUUUCCCGCCCGACCCACUCCG .(((((.(((((((((((....))-)))))))))......((((((..((((.(((......-..(-....)(((((....)))))))).))))..))))))......)))))....... ( -25.60) >DroSec_CAF1 14830 106 + 1 CGGUCGCUAAUUGCCUUUGCCAGAAAGUUAAUUACGGCGAAAAUAAUA---UAUUUGAAAAU--UU-CAAAAUUGGACUUGUCAAAAACAUU-UUGACGUUUCCCCUCAGACG------- ..((((.((((((.((((.....)))).))))))))))((........---..(((((....--.)-))))...(((..(((((((.....)-))))))..)))..)).....------- ( -21.10) >DroMoj_CAF1 19129 93 + 1 CAG-CGCUAAUUGCCUUAGCCC-AAAGUUAAUUACAGCAAA-----------AUUUUCAAAUUUUC-CAAAAUUGGACUUGUCAAAAACAUU-UUGACAUUUCCCACU------------ ..(-(..((((((.(((.....-.))).))))))..))...-----------..............-.......(((..(((((((.....)-))))))..)))....------------ ( -16.20) >DroAna_CAF1 2504 93 + 1 CGGUCGCUAAUUGCCUUUGCCAGAAAGUUAAUUACGGCGAA-----------AUUUGAAAAU--UUCCAAAAUUGGACUUGUCAAAAACAUU-UUGACAUUUCCCC------G------- ..((((.((((((.((((.....)))).))))))))))(((-----------(((....)))--))).......(((..(((((((.....)-))))))..)))..------.------- ( -22.00) >DroPer_CAF1 59732 117 + 1 CGGUCGCUAAUUGGCUUUCCCAGA-AGUUAAUUACGAUGAAAAUGUUGAAAACGUUGAAAAU-GCU-GAAAAUUUGACUUGUCAAAAACAUUUUUGACAUUUCCCGCCCGACCCUCUCCG .(((((.(((((((((((....))-)))))))))......((((((..((((.(((......-..(-....)(((((....)))))))).))))..))))))......)))))....... ( -25.60) >consensus CGGUCGCUAAUUGCCUUUGCCAGAAAGUUAAUUACGGCGAA___________AUUUGAAAAU__UU_CAAAAUUGGACUUGUCAAAAACAUU_UUGACAUUUCCCCCC_GACG_______ ..((((.(((((..((((.....))))..)))))))))....................................(((..((((((........))))))..)))................ (-15.11 = -15.67 + 0.56)

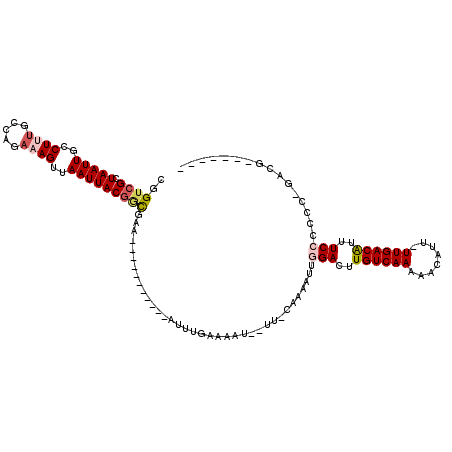
| Location | 18,795,454 – 18,795,545 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.38 |
| Mean single sequence MFE | -15.42 |
| Consensus MFE | -9.95 |
| Energy contribution | -9.82 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18795454 91 + 20766785 A--------AUUUGAAAAUUU-CAAAAUUGGACUUGUCAAAAACAUUUUGACGUUUCCCCUCAGACGUCCGCCAAC-----CACCACCACACGAAAUUCAGCCAU .--------.(((((.....)-))))...((((..((((((.....))))))((((......))))))))......-----........................ ( -13.80) >DroSec_CAF1 14870 95 + 1 AAAUAAUAUAUUUGAAAAUUU-CAAAAUUGGACUUGUCAAAAACAUUUUGACGUUUCCCCUCAGACGUCCGCCACC-----CAGCAC----CGAAAUUCAGCCAU ...........((((..((((-(......((((..((((((.....))))))((((......))))))))((....-----..))..----.))))))))).... ( -17.40) >DroSim_CAF1 14939 87 + 1 A--------AUUUGAAAAUUU-CAAAAUUGGACUUGUCAAAAACAUUUUGACGUUUCCCCUCAGACGUCCGCCACC-----CAGCGC----CGAAAUUCAGCCAU .--------..((((..((((-(......((((..((((((.....))))))((((......))))))))((....-----..))..----.))))))))).... ( -17.40) >DroEre_CAF1 15307 96 + 1 A--------AUUUGAAAAUUU-CAAAAUUGGACUUGUCAAAAACAUUUUGACGUUUCCCCUCAGACAUCCGCCCUCGCCCUCAACAUCACCCGAAAUUCAGCCAU .--------..((((..((((-(......(((..(((((((.....)))))))..)))............((....))..............))))))))).... ( -13.70) >DroYak_CAF1 15130 87 + 1 A--------AUUUGAAAAUUU-CAAAAUUGGACUUGUCAAAAACAUUUUGACGUUUCCCCUCAGACGUCCGCC---------ACCCUCACCCGAAAUUCAGCCAU .--------.(((((.....)-))))...((((..((((((.....))))))((((......))))))))...---------....................... ( -13.80) >DroAna_CAF1 2544 80 + 1 A--------AUUUGAAAAUUUCCAAAAUUGGACUUGUCAAAAACAUUUUGACAUUUCCCC------GCCCGUCACU-----CCGGUU----CG--CUCCGGCCGU .--------.((((........))))...(((..(((((((.....)))))))..)))..------..........-----((((..----..--..)))).... ( -16.40) >consensus A________AUUUGAAAAUUU_CAAAAUUGGACUUGUCAAAAACAUUUUGACGUUUCCCCUCAGACGUCCGCCACC_____CAGCAC____CGAAAUUCAGCCAU .............................(((..(((((((.....)))))))..)))............................................... ( -9.95 = -9.82 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:12 2006