
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,794,792 – 18,794,888 |
| Length | 96 |
| Max. P | 0.848775 |

| Location | 18,794,792 – 18,794,888 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 82.96 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -21.10 |
| Energy contribution | -22.10 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
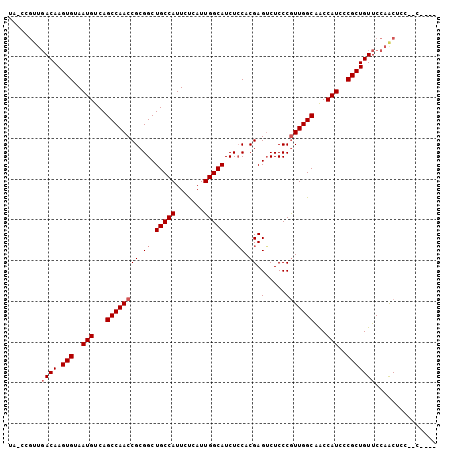
>2R_DroMel_CAF1 18794792 96 + 20766785 UAUCCGUUGACAAGUGUAAUGUCAGCCAACCGCGGCUGCCAUUCUCAUUGGCAUCUGCACGAGUCUCCCAUUGGCAACCAUCCCGCUGUUCCAACU--------- .....((((...((((..(((...(((((..((((.(((((.......))))).))))..(.....)...)))))...)))..))))....)))).--------- ( -28.10) >DroSec_CAF1 14205 96 + 1 ---------ACAAGUGUAAUGUCAGCCAACCGCGGCUGCCAUUCUCAUUGGCAUCUCCACGAGUCUCCCGUUGGCAACCAUCCCGCUGUUCCAACUCCAGCUCCU ---------...((((..(((...((((((((.((.(((((.......)))))...)).))........))))))...)))..)))).................. ( -23.30) >DroEre_CAF1 14629 101 + 1 UACCCGUUGACAAGUGUAAUGUCAGCCAACCGCGGCUGCCAUUCCCAUUGGCAUCUCCACGAGGCUCCCGUUGGCGGCCAUCCCGCUGUUCCAGCUCCUCC---- .....((((...((((..(((.(.((((((.(.((((((((.......))))).(((...)))))).).)))))).).)))..))))....))))......---- ( -31.40) >consensus UA_CCGUUGACAAGUGUAAUGUCAGCCAACCGCGGCUGCCAUUCUCAUUGGCAUCUCCACGAGUCUCCCGUUGGCAACCAUCCCGCUGUUCCAACUCC__C____ ........((((.(((..(((...((((((((.((.(((((.......)))))...)).)).(.....)))))))...)))..)))))))............... (-21.10 = -22.10 + 1.00)


| Location | 18,794,792 – 18,794,888 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 82.96 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -30.61 |
| Energy contribution | -30.50 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
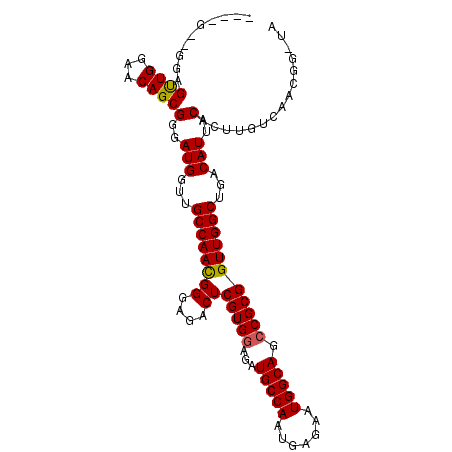
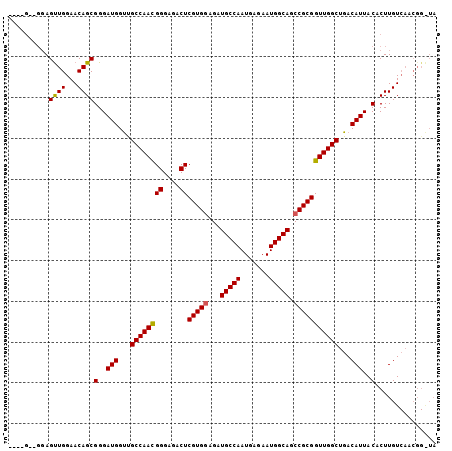
>2R_DroMel_CAF1 18794792 96 - 20766785 ---------AGUUGGAACAGCGGGAUGGUUGCCAAUGGGAGACUCGUGCAGAUGCCAAUGAGAAUGGCAGCCGCGGUUGGCUGACAUUACACUUGUCAACGGAUA ---------.(((((...((.(..(((...((((((((....))((((....(((((.......)))))..))))))))))...)))..).))..)))))..... ( -31.60) >DroSec_CAF1 14205 96 - 1 AGGAGCUGGAGUUGGAACAGCGGGAUGGUUGCCAACGGGAGACUCGUGGAGAUGCCAAUGAGAAUGGCAGCCGCGGUUGGCUGACAUUACACUUGU--------- ....((((.........))))(..(((...((((((((....))(((((...(((((.......))))).)))))))))))...)))..)......--------- ( -37.30) >DroEre_CAF1 14629 101 - 1 ----GGAGGAGCUGGAACAGCGGGAUGGCCGCCAACGGGAGCCUCGUGGAGAUGCCAAUGGGAAUGGCAGCCGCGGUUGGCUGACAUUACACUUGUCAACGGGUA ----......((((...))))......(((((((((((....))(((((...(((((.......))))).)))))))))))(((((.......)))))...))). ( -36.60) >consensus ____G__GGAGUUGGAACAGCGGGAUGGUUGCCAACGGGAGACUCGUGGAGAUGCCAAUGAGAAUGGCAGCCGCGGUUGGCUGACAUUACACUUGUCAACGG_UA ..........((((...))))(..(((...((((((((....))(((((...(((((.......))))).)))))))))))...)))..)............... (-30.61 = -30.50 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:10 2006