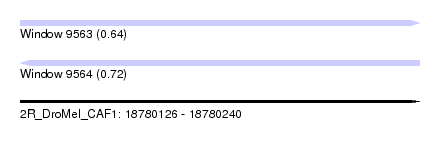
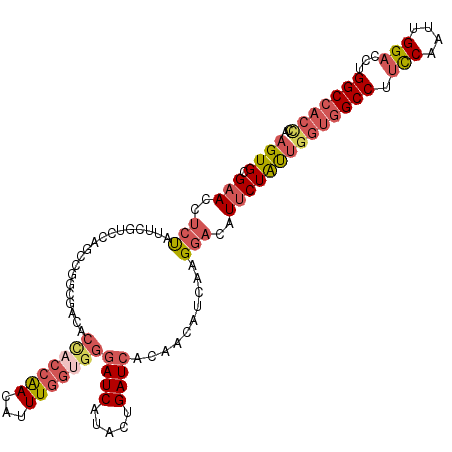
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,780,126 – 18,780,240 |
| Length | 114 |
| Max. P | 0.721330 |

| Location | 18,780,126 – 18,780,240 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.57 |
| Mean single sequence MFE | -41.33 |
| Consensus MFE | -24.08 |
| Energy contribution | -25.75 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.637443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18780126 114 + 20766785 UGGGUGGCCAGGUCCAAUUGGAAGGCCACCAAUAGAAUGUCCUUGAUGUUGUGAUCAGUAUGAUCCCACCAAAUGUUGGAAGUAUCACCGGCUCGACGAAUAGACGUUCGCACU ..(((((((...(((....))).)))))))....(((((((..(..(((((.((((.....)))).........(((((........))))).))))).)..)))))))..... ( -37.90) >DroVir_CAF1 4462 114 + 1 AUAUGUGCCAGUGCCAUUUGAAAGGCCACCAGCAGUAUGUCCAUUAUGUUAUGAUCAGCGCGAUGCCAAAAACAGUCGGUUUCAUCGCGUGUUAACUUGAUGCUGGUGCGCACA ...(((((....(((........)))((((((((...((..(((......)))..))((((((((....((((.....))))))))))))..........)))))))).))))) ( -37.10) >DroSec_CAF1 41 114 + 1 UGGGUGGCCAGGUCCAAUUGGAAGGCCACCAAUAGAAUGUCCUUGAUGUUGUGAUCAGUAUGAUCCCACCAAAUGUUGGUGGUGUCACCGGCUGGACGAAUAGACGUUCGCACU ..(((((((...(((....))).)))))))....(((((((.......((((..(((((.(((..((((((.....))))))..)))...)))))))))...)))))))..... ( -46.40) >DroSim_CAF1 41 114 + 1 UGGGUGGCCAGGUCCAAUUGGAAGGCCACCAAUAGAAUGUCCUUGAUGUUGUGAUCAGUAUGAUCCCACCAAAUGUUGGUGGUGUCACCGGCUGGACGAAUAGACGUUCGCACU ..(((((((...(((....))).)))))))....(((((((.......((((..(((((.(((..((((((.....))))))..)))...)))))))))...)))))))..... ( -46.40) >DroEre_CAF1 41 114 + 1 UGGGUGGCCAGGUCCAAUUGAAAGGCCACCAAUAGAAUAUCCUUGAUGUUGUGAUCAGUAUGAUCCCACCAAAUGUUGGUGGUGUCGCCGGCUGGUCGAAUGGAGGUUCGCACU ..(((((((...((.....))..)))))))....(((...((((.((....((((((((.(((..((((((.....))))))..)))...)))))))).)).)))))))..... ( -44.60) >DroYak_CAF1 41 114 + 1 UGCGUGGCCAGGUCCAACUGAAAGGCCACCAGUAGAAUAUCCUUGAUGUUGUGAUCGGUAUGAUCCCUCCAAAUGUUGGUGGUAUCGCCGGCUGGUCGAAUAGAGGUUCUCACU ((.((((((...((.....))..))))))))((((((...((((..((((.((((((((.((((.((.(((.....))).)).))))...)))))))))))))))))))).)). ( -35.60) >consensus UGGGUGGCCAGGUCCAAUUGAAAGGCCACCAAUAGAAUGUCCUUGAUGUUGUGAUCAGUAUGAUCCCACCAAAUGUUGGUGGUAUCACCGGCUGGACGAAUAGACGUUCGCACU ..(((((((...((.....))..)))))))..((.((((((...)))))).)).(((((.((((.((((((.....)))))).))))...))))).(((((....))))).... (-24.08 = -25.75 + 1.67)

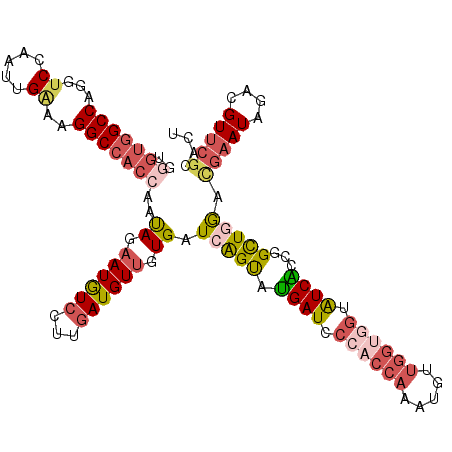
| Location | 18,780,126 – 18,780,240 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 82.57 |
| Mean single sequence MFE | -36.97 |
| Consensus MFE | -22.57 |
| Energy contribution | -24.05 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18780126 114 - 20766785 AGUGCGAACGUCUAUUCGUCGAGCCGGUGAUACUUCCAACAUUUGGUGGGAUCAUACUGAUCACAACAUCAAGGACAUUCUAUUGGUGGCCUUCCAAUUGGACCUGGCCACCCA .....(((.((((..((..((...))..))............((((((.((((.....))))....)))))))))).)))....(((((((.(((....)))...))))))).. ( -35.20) >DroVir_CAF1 4462 114 - 1 UGUGCGCACCAGCAUCAAGUUAACACGCGAUGAAACCGACUGUUUUUGGCAUCGCGCUGAUCAUAACAUAAUGGACAUACUGCUGGUGGCCUUUCAAAUGGCACUGGCACAUAU (((((.((((((((....(((..(((((((((...((((......))))))))))).))..(((......))))))....))))))))(((........)))....)))))... ( -40.30) >DroSec_CAF1 41 114 - 1 AGUGCGAACGUCUAUUCGUCCAGCCGGUGACACCACCAACAUUUGGUGGGAUCAUACUGAUCACAACAUCAAGGACAUUCUAUUGGUGGCCUUCCAAUUGGACCUGGCCACCCA .................((((....((((...((((((.....))))))((((.....))))....))))..))))........(((((((.(((....)))...))))))).. ( -39.80) >DroSim_CAF1 41 114 - 1 AGUGCGAACGUCUAUUCGUCCAGCCGGUGACACCACCAACAUUUGGUGGGAUCAUACUGAUCACAACAUCAAGGACAUUCUAUUGGUGGCCUUCCAAUUGGACCUGGCCACCCA .................((((....((((...((((((.....))))))((((.....))))....))))..))))........(((((((.(((....)))...))))))).. ( -39.80) >DroEre_CAF1 41 114 - 1 AGUGCGAACCUCCAUUCGACCAGCCGGCGACACCACCAACAUUUGGUGGGAUCAUACUGAUCACAACAUCAAGGAUAUUCUAUUGGUGGCCUUUCAAUUGGACCUGGCCACCCA .....(((..(((..(((.((....)))))..((((((.....))))))((((.....))))..........)))..)))....(((((((.(((....)))...))))))).. ( -37.00) >DroYak_CAF1 41 114 - 1 AGUGAGAACCUCUAUUCGACCAGCCGGCGAUACCACCAACAUUUGGAGGGAUCAUACCGAUCACAACAUCAAGGAUAUUCUACUGGUGGCCUUUCAGUUGGACCUGGCCACGCA (((.((((..(((..(((.((....)))))..((.(((.....))).))((((.....))))..........)))..))))))).((((((.(((....)))...))))))... ( -29.70) >consensus AGUGCGAACCUCUAUUCGUCCAGCCGGCGACACCACCAACAUUUGGUGGGAUCAUACUGAUCACAACAUCAAGGACAUUCUAUUGGUGGCCUUCCAAUUGGACCUGGCCACCCA ((((.(((..(((...................(((((((...)))))))((((.....))))..........)))..)))))))(((((((.(((....)))...))))))).. (-22.57 = -24.05 + 1.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:08 2006