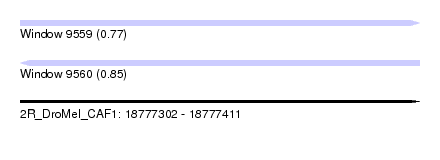
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,777,302 – 18,777,411 |
| Length | 109 |
| Max. P | 0.845507 |

| Location | 18,777,302 – 18,777,411 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.20 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -18.82 |
| Energy contribution | -18.98 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
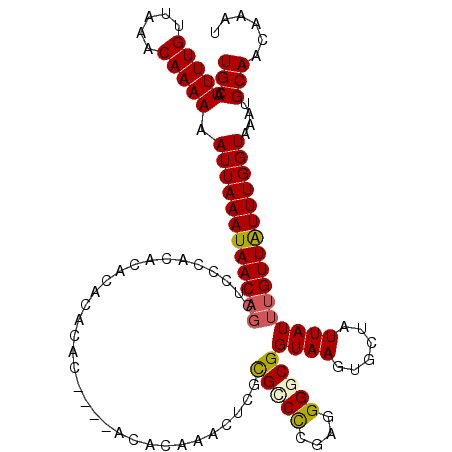
>2R_DroMel_CAF1 18777302 109 + 20766785 UGCAUUUUGUUAAGCAAAAAAUUAAAUAACAGCACCCACACACAC--------ACACACACUCGUGGCCAGAGGGGCGGUAAGUGCUAUUAUUUGUUAUUUGGUAAACGCAACAAAU (((.(((((.....))))).(((((((((((((((((........--------.(((......)))(((.....)))))...)))).......)))))))))))....)))...... ( -24.51) >DroSec_CAF1 140039 113 + 1 UGCAUUUUGUUAAGCAAAAAAUUAAAUAACAGCUCCCACACACACAAACACA----CACACUCGCGCCCAGAGGGGCGGUAAGUGCUAUUAUUUGUUAUUUGGUAAACGCAACAAAU (((.(((((.....))))).((((((((((((......).............----..((((..(((((....)))).)..))))........)))))))))))....)))...... ( -24.70) >DroEre_CAF1 140954 113 + 1 UGCAUUUUGUUAAACAAAAAAUUAAAUAACAGCCCCCGCACACACACAC----ACACAAAUACGCGGCCCGAGGGGCGGUAAGUGCUAUUAUUUGUUGUUUGGUAAAUGCAACAAAU (((((((.(..((((((.....(((((((.(((...(((..........----..........)))((((...)))).......))).)))))))))))))..))))))))...... ( -24.95) >DroYak_CAF1 142518 100 + 1 UGCAUUUUGUUAAACAAAAAAUUAAAUAAC-------------ACACAC----AUACAGACUCGCGUCCCGAGGGGCGGUAAGUGCUAUUAUUUGUUGUUUGGUAAAUGCAACAAAU (((((((.(..((((((.((((..((((.(-------------((...(----.....).....(((((....)))))....))).)))))))).))))))..))))))))...... ( -23.40) >DroAna_CAF1 139341 114 + 1 UGCAUUUUGUUGAACAAAAAAUUAAAUAACAGCUCCCACACACACGC-CGCAUGCCGCAGGACCCGCCUCGAGGGGCGGUAAGUGCUAUUAUUUGUUAUUUGGUAAAUGCAACA--U (((((((((.....))))).((((((((((((......(((.....(-(((.....)).))..((((((....))))))...))).......))))))))))))...))))...--. ( -30.42) >consensus UGCAUUUUGUUAAACAAAAAAUUAAAUAACAGCUCCCACACACACACAC____ACACAAACUCGCGCCCCGAGGGGCGGUAAGUGCUAUUAUUUGUUAUUUGGUAAAUGCAACAAAU (((.(((((.....))))).((((((((((((................................(((((....)))))((((......))))))))))))))))....)))...... (-18.82 = -18.98 + 0.16)


| Location | 18,777,302 – 18,777,411 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.20 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -13.10 |
| Energy contribution | -15.86 |
| Covariance contribution | 2.76 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
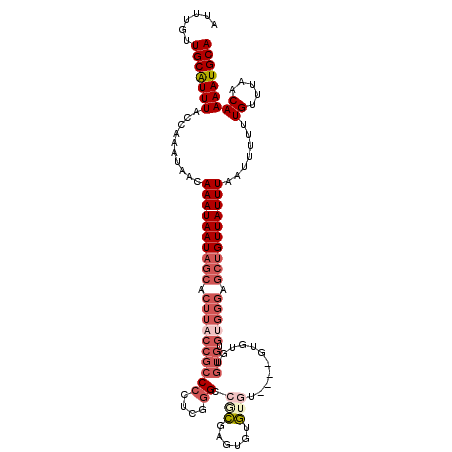
>2R_DroMel_CAF1 18777302 109 - 20766785 AUUUGUUGCGUUUACCAAAUAACAAAUAAUAGCACUUACCGCCCCUCUGGCCACGAGUGUGUGU--------GUGUGUGUGGGUGCUGUUAUUUAAUUUUUUGCUUAACAAAAUGCA .(((((((((((........)))((((((((((((((((((((.....)))((((......)))--------)...).))))))))))))))))........))..))))))..... ( -29.80) >DroSec_CAF1 140039 113 - 1 AUUUGUUGCGUUUACCAAAUAACAAAUAAUAGCACUUACCGCCCCUCUGGGCGCGAGUGUG----UGUGUUUGUGUGUGUGGGAGCUGUUAUUUAAUUUUUUGCUUAACAAAAUGCA ......(((((((.(((....(((((((...((((((..(((((....))))).)))))).----..))))))).....)))....(((((..(((....)))..)))))))))))) ( -35.80) >DroEre_CAF1 140954 113 - 1 AUUUGUUGCAUUUACCAAACAACAAAUAAUAGCACUUACCGCCCCUCGGGCCGCGUAUUUGUGU----GUGUGUGUGUGCGGGGGCUGUUAUUUAAUUUUUUGUUUAACAAAAUGCA ......(((((((...((((((.(((((((((((......(((((.((.(((((((((......----))))))).)).))))))))))))))..)))).))))))....))))))) ( -34.10) >DroYak_CAF1 142518 100 - 1 AUUUGUUGCAUUUACCAAACAACAAAUAAUAGCACUUACCGCCCCUCGGGACGCGAGUCUGUAU----GUGUGU-------------GUUAUUUAAUUUUUUGUUUAACAAAAUGCA ......(((((((...((((((.((((((((((((...(((.....)))(((....))).....----....))-------------))))))..)))).))))))....))))))) ( -25.10) >DroAna_CAF1 139341 114 - 1 A--UGUUGCAUUUACCAAAUAACAAAUAAUAGCACUUACCGCCCCUCGAGGCGGGUCCUGCGGCAUGCG-GCGUGUGUGUGGGAGCUGUUAUUUAAUUUUUUGUUCAACAAAAUGCA .--...(((((((..........((((((((((.....(((((......))))).(((..((.(((((.-..)))))))..))))))))))))).......((.....))))))))) ( -36.30) >consensus AUUUGUUGCAUUUACCAAAUAACAAAUAAUAGCACUUACCGCCCCUCGGGCCGCGAGUGUGUGU____GUGUGUGUGUGUGGGAGCUGUUAUUUAAUUUUUUGUUUAACAAAAUGCA ......(((((((..........((((((((((.((((((((((....)).(((......)))...........))).))))).)))))))))).......((.....))))))))) (-13.10 = -15.86 + 2.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:03 2006