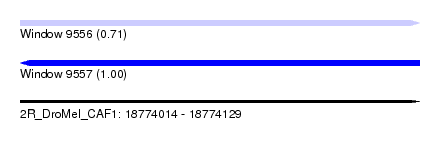
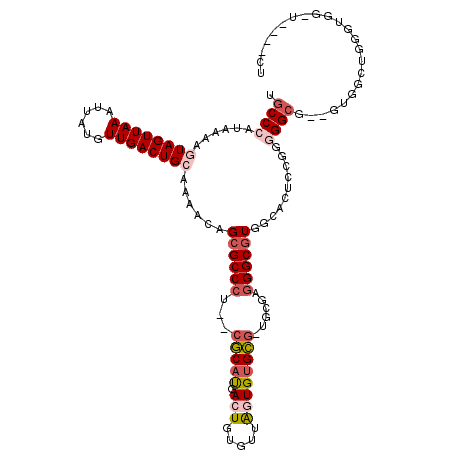
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,774,014 – 18,774,129 |
| Length | 115 |
| Max. P | 0.995331 |

| Location | 18,774,014 – 18,774,129 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.46 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -15.32 |
| Energy contribution | -15.64 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18774014 115 + 20766785 AG-ACCACCCACCCAGCCACUCCGCCGCCGGAGUGCCACGCCCACGUA-CGCACACUAACACAGUCAAGCG--AGGGCGCUGUUUUGCAGUCAACAUAAUUUAACUACUUUUAUGGGCA ..-........(((((.(((((((....))))))).)..((((.(((.-.((...........))...)))--.))))((((.....))))......................)))).. ( -29.70) >DroSec_CAF1 136778 114 + 1 AGGAAAAGCCACCCAGCCAC--CGCCCCCGGAGUGCCACGCCCACGUA-CGCACACUAACACAGUCAUGCG--AGGGCGCUGUUUUGCAGUCAACAUAAUUUAACUACUUUUAUGGGCA .((....((......))..)--)((((..((((((....((((.((((-.(.((.........))).))))--.))))((((.....))))..............))))))...)))). ( -28.30) >DroEre_CAF1 137860 100 + 1 --------------GCCAAC--CGCCCCCGCAGUGCCACGCCCUCGCA-CGCACACCAACACACUCAUGCG--AGGGCGCUGUUUUUCAGUCAACAUAAUUUAACUACUUUUAUGGGCA --------------......--.((((....((((....(((((((((-..................))))--)))))((((.....))))..............)))).....)))). ( -29.07) >DroYak_CAF1 139069 108 + 1 AA-CCC-----ACCACCCAC--CGCCACCGCAGUGCCACGCCCUCGCA-CGCACACUAACACACUCAUGCG--AGGGCGCUGUUUUGCAGUCAACAUAAUUUAACUACUUUUAUGGGCA ..-...-----.........--.(((...((((.((..((((((((((-..................))))--))))))..)).))))......(((((...........)))))))). ( -32.97) >DroAna_CAF1 136702 111 + 1 UGU---AACCAACAUGCCAC--GCCCCCC---GCCCUAUGCCUCCUCUGGCCACACUCACACAGUCGUGGAUACUGGCGCUGUUUUUCAGUCAACAUAAUUUAACUACUUUUAUGGGCA ...---..............--((((...---(((....(((......)))((((((.....))).)))......)))((((.....)))).......................)))). ( -22.70) >consensus AG____A_CCAACCAGCCAC__CGCCCCCGCAGUGCCACGCCCACGCA_CGCACACUAACACAGUCAUGCG__AGGGCGCUGUUUUGCAGUCAACAUAAUUUAACUACUUUUAUGGGCA ..................................(((..((((......((((..............))))...))))((((.....))))...(((((...........)))))))). (-15.32 = -15.64 + 0.32)

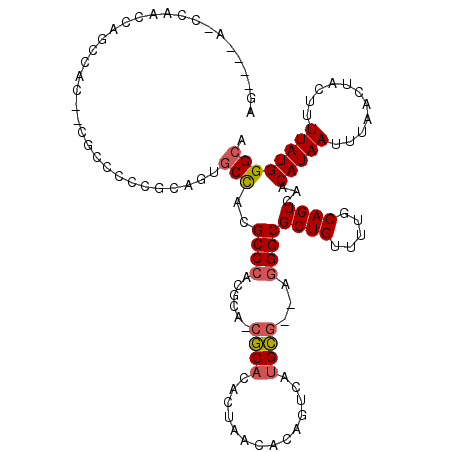
| Location | 18,774,014 – 18,774,129 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.46 |
| Mean single sequence MFE | -43.14 |
| Consensus MFE | -22.60 |
| Energy contribution | -24.16 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18774014 115 - 20766785 UGCCCAUAAAAGUAGUUAAAUUAUGUUGACUGCAAAACAGCGCCCU--CGCUUGACUGUGUUAGUGUGCG-UACGUGGGCGUGGCACUCCGGCGGCGGAGUGGCUGGGUGGGUGGU-CU .((((((....((((((((......))))))))....((((((((.--(((...((((...))))..)))-.....))))....(((((((....))))))))))).))))))...-.. ( -47.10) >DroSec_CAF1 136778 114 - 1 UGCCCAUAAAAGUAGUUAAAUUAUGUUGACUGCAAAACAGCGCCCU--CGCAUGACUGUGUUAGUGUGCG-UACGUGGGCGUGGCACUCCGGGGGCG--GUGGCUGGGUGGCUUUUCCU .(((((.....((((((((......))))))))....((.((((((--(((((....))))..(.((((.-((((....)))))))).).)))))))--.))..))))).......... ( -43.80) >DroEre_CAF1 137860 100 - 1 UGCCCAUAAAAGUAGUUAAAUUAUGUUGACUGAAAAACAGCGCCCU--CGCAUGAGUGUGUUGGUGUGCG-UGCGAGGGCGUGGCACUGCGGGGGCG--GUUGGC-------------- .((((......(((((.......((((........))))(((((((--((((((.(..(....)..).))-)))))))))))...)))))..)))).--......-------------- ( -40.60) >DroYak_CAF1 139069 108 - 1 UGCCCAUAAAAGUAGUUAAAUUAUGUUGACUGCAAAACAGCGCCCU--CGCAUGAGUGUGUUAGUGUGCG-UGCGAGGGCGUGGCACUGCGGUGGCG--GUGGGUGGU-----GGG-UU .((((((....((((((((......))))))))...((.(((((((--((((((.(..(....)..).))-)))))))))))..((((((....)))--))).)).))-----)))-). ( -51.50) >DroAna_CAF1 136702 111 - 1 UGCCCAUAAAAGUAGUUAAAUUAUGUUGACUGAAAAACAGCGCCAGUAUCCACGACUGUGUGAGUGUGGCCAGAGGAGGCAUAGGGC---GGGGGGC--GUGGCAUGUUGGUU---ACA .((((.......(((((((......)))))))........((((......(((.(((.....))))))(((......)))....)))---)..))))--(((((......)))---)). ( -32.70) >consensus UGCCCAUAAAAGUAGUUAAAUUAUGUUGACUGCAAAACAGCGCCCU__CGCAUGACUGUGUUAGUGUGCG_UGCGAGGGCGUGGCACUCCGGGGGCG__GUGGCUGGGUGG_U____CU .((((......((((((((......))))))))......((((((...(((((.(((.....))))))))......))))))..........))))....................... (-22.60 = -24.16 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:01 2006