
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 3,033,588 – 3,033,692 |
| Length | 104 |
| Max. P | 0.930770 |

| Location | 3,033,588 – 3,033,692 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.65 |
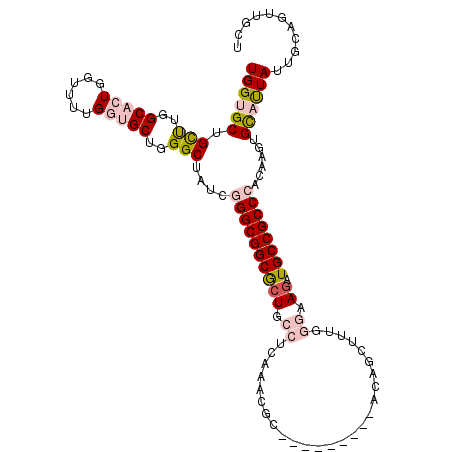
| Mean single sequence MFE | -42.77 |
| Consensus MFE | -29.52 |
| Energy contribution | -30.52 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
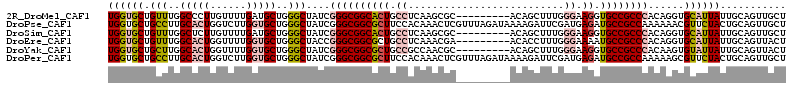
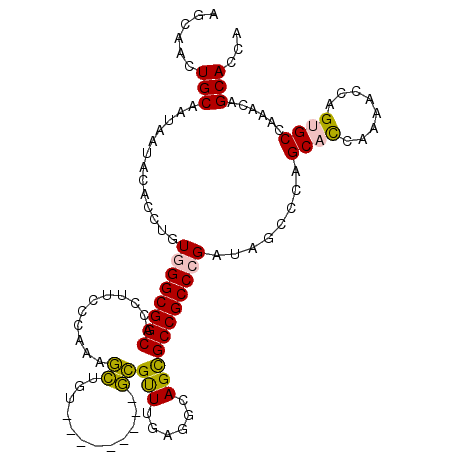
>2R_DroMel_CAF1 3033588 104 + 20766785 UGGUGCUGUUUGGCCCUUGUUUUGAUGCUGGGCUAUCGGGCGGCACUGCCUCAAGCGC---------ACAGCUUUGGGAAGGUGCCGCCCACAGGUGCAUUAUUGCAGUUGCU ..........((((((..((......)).))))))..((((((((((.(((.((((..---------...)))).)))..)))))))))).(((.((((....)))).))).. ( -48.90) >DroPse_CAF1 10492 113 + 1 UGGUGCUGCCUUGCACUGGUCUUGGUGCUGGGCUAUCGGGCGGCGCUUCCACAAACUCGUUUAGAUAAAAGAUUCGAUGAGAUGCCGCCAAAAAACGUUCUACUGCAGUUGCU ...(((.((((.(((((......))))).)))).....(((((((......)...((((((..((........))))))))..))))))...............)))...... ( -33.70) >DroSim_CAF1 7232 104 + 1 UGGUGCUGUUUGGCUCUUGUUUUGAUGCUGGGCUAUCGGGCGGCACUGCCUCAAGCGC---------ACAGCUUUGGGAAGGUGCCGCCCACAGGUGCAUUAUUGCAGUUGCU .((((..(((..((((.......)).))..)))))))((((((((((.(((.((((..---------...)))).)))..)))))))))).(((.((((....)))).))).. ( -47.80) >DroEre_CAF1 7680 104 + 1 UGGUGCUGUUUGGCACUGGUUUUGGUGCUGGGCUACCGGGCGGCGCUGCCUCAAACGA---------ACACCUUUGGGAAAAUGCCGCCCACAGGUGCAUUAUUGCAGUUACU .((((..(((..(((((......)))))..)))))))((((((((.(..((((((...---------.....))))))..).))))))))..((.((((....)))).))... ( -43.20) >DroYak_CAF1 7356 104 + 1 UGGUGCUGCUUGGCACUGGUUUUGGUGCUGGGCUAUCGGGCGGCGCUGCCGCCAACGC---------ACAGCUUUGGGAAGGUGCCGCCCACAAGUGUAUUAUUGCAGUUACU .((((..(((..(((((......)))))..)))))))((((((((((..(.((((.((---------...)).)))))..))))))))))..((.((((....)))).))... ( -48.00) >DroPer_CAF1 10274 113 + 1 UGGUGCUGCCUUGCACUGGUCUUGGUGCUGGGCUAUCGGGCGGCGCUUCCACAAACUCGUUUAGAUAAAAGAUUCGAUGAGAUGCCGCCAAAAAGCGUUCUACUGCAGUUGCU ....(((((((.(((((......))))).)))).....(((((((......)...((((((..((........))))))))..))))))....)))........((....)). ( -35.00) >consensus UGGUGCUGCUUGGCACUGGUUUUGGUGCUGGGCUAUCGGGCGGCGCUGCCUCAAACGC_________ACAGCUUUGGGAAGAUGCCGCCCACAAGUGCAUUAUUGCAGUUGCU ((((((.(((..(((((......)))))..)))....((((((((((.((..........................)).)).))))))))......))))))........... (-29.52 = -30.52 + 1.00)
| Location | 3,033,588 – 3,033,692 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.65 |
| Mean single sequence MFE | -33.55 |
| Consensus MFE | -21.50 |
| Energy contribution | -20.78 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
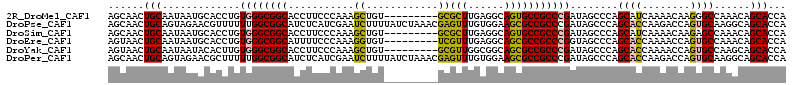
>2R_DroMel_CAF1 3033588 104 - 20766785 AGCAACUGCAAUAAUGCACCUGUGGGCGGCACCUUCCCAAAGCUGU---------GCGCUUGAGGCAGUGCCGCCCGAUAGCCCAGCAUCAAAACAAGGGCCAAACAGCACCA .((...((((....))))..((((((((((((...((..((((...---------..))))..))..)))))))))....((((.............))))...))))).... ( -38.72) >DroPse_CAF1 10492 113 - 1 AGCAACUGCAGUAGAACGUUUUUUGGCGGCAUCUCAUCGAAUCUUUUAUCUAAACGAGUUUGUGGAAGCGCCGCCCGAUAGCCCAGCACCAAGACCAGUGCAAGGCAGCACCA ......(((...............((((((.(..((.((((.(((..........)))))))))..)..)))))).....(((..((((........))))..))).)))... ( -29.20) >DroSim_CAF1 7232 104 - 1 AGCAACUGCAAUAAUGCACCUGUGGGCGGCACCUUCCCAAAGCUGU---------GCGCUUGAGGCAGUGCCGCCCGAUAGCCCAGCAUCAAAACAAGAGCCAAACAGCACCA .((...((((....)))).(((((((((((((...((..((((...---------..))))..))..))))))))).))))....))............((......)).... ( -35.10) >DroEre_CAF1 7680 104 - 1 AGUAACUGCAAUAAUGCACCUGUGGGCGGCAUUUUCCCAAAGGUGU---------UCGUUUGAGGCAGCGCCGCCCGGUAGCCCAGCACCAAAACCAGUGCCAAACAGCACCA ......(((......((((..(((((((((.....((((((.(...---------.).)))).))....)))))))(((.((...)))))...))..))))......)))... ( -32.50) >DroYak_CAF1 7356 104 - 1 AGUAACUGCAAUAAUACACUUGUGGGCGGCACCUUCCCAAAGCUGU---------GCGUUGGCGGCAGCGCCGCCCGAUAGCCCAGCACCAAAACCAGUGCCAAGCAGCACCA .....((((.............(((((.(((((((....)))..))---------))...((((((...)))))).....)))))((((........))))...))))..... ( -34.90) >DroPer_CAF1 10274 113 - 1 AGCAACUGCAGUAGAACGCUUUUUGGCGGCAUCUCAUCGAAUCUUUUAUCUAAACGAGUUUGUGGAAGCGCCGCCCGAUAGCCCAGCACCAAGACCAGUGCAAGGCAGCACCA .....((((........(((.((.((((((.(..((.((((.(((..........)))))))))..)..)))))).)).)))...((((........))))...))))..... ( -30.90) >consensus AGCAACUGCAAUAAUACACCUGUGGGCGGCACCUUCCCAAAGCUGU_________GCGUUUGAGGCAGCGCCGCCCGAUAGCCCAGCACCAAAACCAGUGCCAAACAGCACCA ......(((.............((((((((...........((............))(((......)))))))))))........((((........))))......)))... (-21.50 = -20.78 + -0.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:23 2006