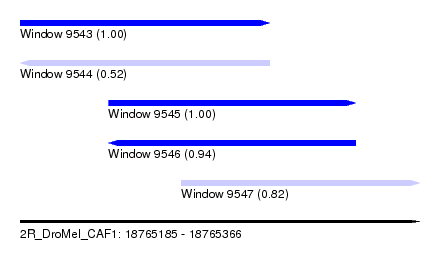
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,765,185 – 18,765,366 |
| Length | 181 |
| Max. P | 0.998226 |

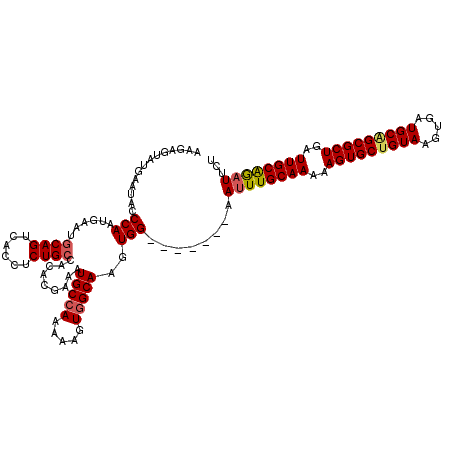
| Location | 18,765,185 – 18,765,298 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.49 |
| Mean single sequence MFE | -36.32 |
| Consensus MFE | -29.11 |
| Energy contribution | -29.62 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.80 |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.998226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18765185 113 + 20766785 AAGAGUAUGAAUACCCAAUGAACGCAGUCACCUCUGCCACACGAAAUGCCAAAAAGUGGCAAGUGG-------AAUUUGCAAAAAGUGCUGUAAGUGAUGCAGCGCUGAUUGCAGAUUCU .......................((((......))))..(((....(((((.....))))).)))(-------(((((((((..(((((((((.....)))))))))..)))))))))). ( -39.70) >DroSec_CAF1 127704 113 + 1 AAGCGUAUGAAAACCCAAUGAAUGCAGUCACCUCUGCCACACGAAAUGCAAAAAACUGGCAAGUGG-------AAUUUGCAAAAAGUGCUGUAAGUGAUGCAGCGCUGAUUGCAGAUUCU ..((((.((......))......((((......))))........)))).....(((....)))((-------(((((((((..(((((((((.....)))))))))..))))))))))) ( -34.60) >DroSim_CAF1 128560 113 + 1 AGGCGUAUGAAUACCCAAUGAAUGCAGUCACCUCUGCCACACGAAAUGCCAAAAAGUGGCAAGUGG-------AAUUUGCAAAAAGUGCUGUAAGUGAUGCAGCGCUGAUUGCAGAUUCU .((.(((....))))).......((((......))))..(((....(((((.....))))).)))(-------(((((((((..(((((((((.....)))))))))..)))))))))). ( -42.30) >DroEre_CAF1 129836 112 + 1 GAAAGUAUGAAUAGCCAAUGAAUGCAGUCACCUCUGGCCGACGAGAUGCCAAAAAGUGGCAAGUGG--------AAUUGCAAAAAGUGCUGUAAGUGAUGCGGCGCUGAUUGCGAAUUCU ....((((..((.....))..)))).(((((...((((.........))))....)))))....((--------((((((((..(((((((((.....)))))))))..)))).)))))) ( -33.00) >DroYak_CAF1 130643 108 + 1 AGAAGUGUGAAUAUCCAAUGAAUGCAGUCACCUCUGCCUGACGAAAUGCCAAAAAGUGGCAAGUGGCAAGUGGCAUUUGCAAAAAGUGCU------------GCGCUGAUUGCAGAUUCU ........((((..........((((((((.(..((((..((....(((((.....))))).)))))).(..((((((.....)))))).------------.)).)))))))).)))). ( -32.00) >consensus AAGAGUAUGAAUACCCAAUGAAUGCAGUCACCUCUGCCACACGAAAUGCCAAAAAGUGGCAAGUGG_______AAUUUGCAAAAAGUGCUGUAAGUGAUGCAGCGCUGAUUGCAGAUUCU ..............(((......((((......)))).........(((((.....)))))..)))........((((((((..(((((((((.....)))))))))..))))))))... (-29.11 = -29.62 + 0.51)

| Location | 18,765,185 – 18,765,298 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.49 |
| Mean single sequence MFE | -28.64 |
| Consensus MFE | -20.17 |
| Energy contribution | -21.22 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18765185 113 - 20766785 AGAAUCUGCAAUCAGCGCUGCAUCACUUACAGCACUUUUUGCAAAUU-------CCACUUGCCACUUUUUGGCAUUUCGUGUGGCAGAGGUGACUGCGUUCAUUGGGUAUUCAUACUCUU .((((.(((((..((.((((.........)))).))..))))).)))-------)....((((((...............))))))(((((((.(((.(.....).))).)))).))).. ( -31.46) >DroSec_CAF1 127704 113 - 1 AGAAUCUGCAAUCAGCGCUGCAUCACUUACAGCACUUUUUGCAAAUU-------CCACUUGCCAGUUUUUUGCAUUUCGUGUGGCAGAGGUGACUGCAUUCAUUGGGUUUUCAUACGCUU .(((..((((.(((.(.((((..........((((....((((((..-------..(((....)))..))))))....)))).)))).).))).)))))))...(.((......)).).. ( -25.90) >DroSim_CAF1 128560 113 - 1 AGAAUCUGCAAUCAGCGCUGCAUCACUUACAGCACUUUUUGCAAAUU-------CCACUUGCCACUUUUUGGCAUUUCGUGUGGCAGAGGUGACUGCAUUCAUUGGGUAUUCAUACGCCU .((((.(((((..((.((((.........)))).))..))))).)))-------)....(((((.....)))))...((((((((((......))))(((.....)))...))))))... ( -31.00) >DroEre_CAF1 129836 112 - 1 AGAAUUCGCAAUCAGCGCCGCAUCACUUACAGCACUUUUUGCAAUU--------CCACUUGCCACUUUUUGGCAUCUCGUCGGCCAGAGGUGACUGCAUUCAUUGGCUAUUCAUACUUUC .((((..((.....))((((.........((((((((((.((....--------.....(((((.....)))))........)).))))))).))).......)))).))))........ ( -25.72) >DroYak_CAF1 130643 108 - 1 AGAAUCUGCAAUCAGCGC------------AGCACUUUUUGCAAAUGCCACUUGCCACUUGCCACUUUUUGGCAUUUCGUCAGGCAGAGGUGACUGCAUUCAUUGGAUAUUCACACUUCU .((((.(.((((.((.((------------((((((((..((....))....((((((.(((((.....)))))....))..)))))))))).)))).)).)))).).))))........ ( -29.10) >consensus AGAAUCUGCAAUCAGCGCUGCAUCACUUACAGCACUUUUUGCAAAUU_______CCACUUGCCACUUUUUGGCAUUUCGUGUGGCAGAGGUGACUGCAUUCAUUGGGUAUUCAUACUCUU .((((..((((..((.((((.........)))).))..))))............(((..(((((.....))))).........((((......))))......)))..))))........ (-20.17 = -21.22 + 1.05)

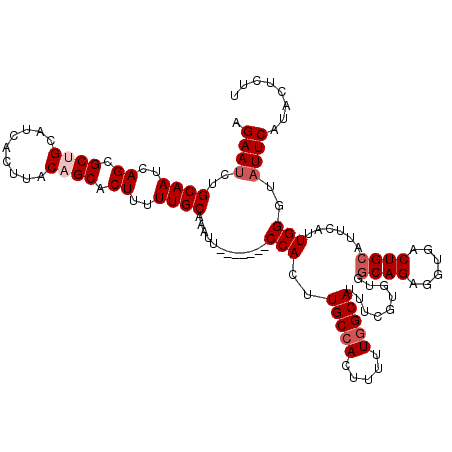
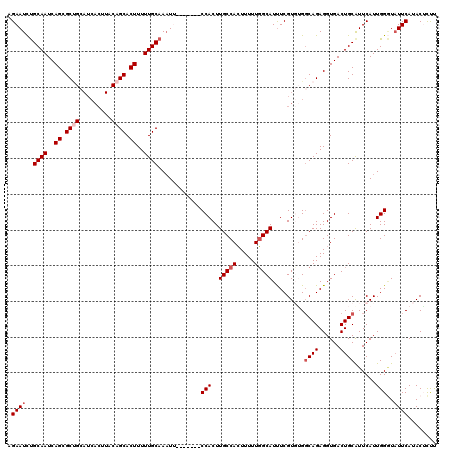
| Location | 18,765,225 – 18,765,337 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 89.11 |
| Mean single sequence MFE | -43.06 |
| Consensus MFE | -34.14 |
| Energy contribution | -35.45 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -5.44 |
| Structure conservation index | 0.79 |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.998102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18765225 112 + 20766785 ACGAAAUGCCAAAAAGUGGCAAGUGG-------AAUUUGCAAAAAGUGCUGUAAGUGAUGCAGCGCUGAUUGCAGAUUCUGUUUCUGCCACAGUGGGGCAAUUAGAAGAAUGCGCCAAA ...............((((((((..(-------(((((((((..(((((((((.....)))))))))..))))))))))..))..))))))..(((.(((.((....)).))).))).. ( -47.40) >DroSec_CAF1 127744 112 + 1 ACGAAAUGCAAAAAACUGGCAAGUGG-------AAUUUGCAAAAAGUGCUGUAAGUGAUGCAGCGCUGAUUGCAGAUUCUGUUUAUGCCACAGUGGGGCAAUUAGAAGAAUGCGCCAAA ................(((((((..(-------(((((((((..(((((((((.....)))))))))..))))))))))..))..)))))...(((.(((.((....)).))).))).. ( -43.40) >DroSim_CAF1 128600 112 + 1 ACGAAAUGCCAAAAAGUGGCAAGUGG-------AAUUUGCAAAAAGUGCUGUAAGUGAUGCAGCGCUGAUUGCAGAUUCUGUUUCUGCCACAGUGGGGCAAUUAGAAGAAUGCGCCAAA ...............((((((((..(-------(((((((((..(((((((((.....)))))))))..))))))))))..))..))))))..(((.(((.((....)).))).))).. ( -47.40) >DroEre_CAF1 129876 111 + 1 ACGAGAUGCCAAAAAGUGGCAAGUGG--------AAUUGCAAAAAGUGCUGUAAGUGAUGCGGCGCUGAUUGCGAAUUCUGUUUCUGCCACAGUUGGGCAAUUAGAAGAAUGCGCCAAA ......((((.((..((((((((..(--------((((((((..(((((((((.....)))))))))..)))).)))))..))..))))))..)).))))................... ( -40.80) >DroYak_CAF1 130683 107 + 1 ACGAAAUGCCAAAAAGUGGCAAGUGGCAAGUGGCAUUUGCAAAAAGUGCU------------GCGCUGAUUGCAGAUUCUGUUUCUGCCACAGCCGGGCAAUUAGAAGAAUGCGCCAAA ......(((((.....)))))..((((..(..((((((.....)))))).------------.)((((...(((((.......)))))..))))...(((.((....)).))))))).. ( -36.30) >consensus ACGAAAUGCCAAAAAGUGGCAAGUGG_______AAUUUGCAAAAAGUGCUGUAAGUGAUGCAGCGCUGAUUGCAGAUUCUGUUUCUGCCACAGUGGGGCAAUUAGAAGAAUGCGCCAAA ......(((((.....))))).((((.......(((((((((..(((((((((.....)))))))))..))))))))).........))))..(((.(((.((....)).))).))).. (-34.14 = -35.45 + 1.31)


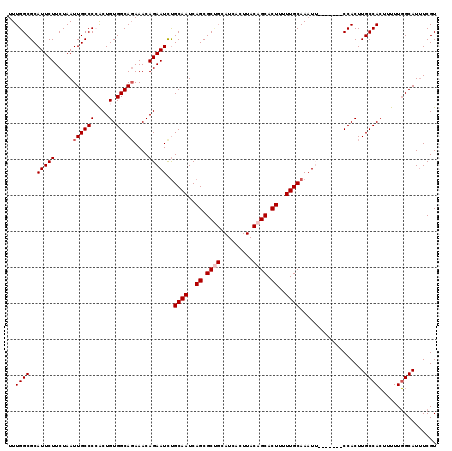
| Location | 18,765,225 – 18,765,337 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 89.11 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -21.17 |
| Energy contribution | -21.82 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18765225 112 - 20766785 UUUGGCGCAUUCUUCUAAUUGCCCCACUGUGGCAGAAACAGAAUCUGCAAUCAGCGCUGCAUCACUUACAGCACUUUUUGCAAAUU-------CCACUUGCCACUUUUUGGCAUUUCGU ...................((((.....(((((((.....((((.(((((..((.((((.........)))).))..))))).)))-------)...))))))).....))))...... ( -31.30) >DroSec_CAF1 127744 112 - 1 UUUGGCGCAUUCUUCUAAUUGCCCCACUGUGGCAUAAACAGAAUCUGCAAUCAGCGCUGCAUCACUUACAGCACUUUUUGCAAAUU-------CCACUUGCCAGUUUUUUGCAUUUCGU ..(((.(((..........))).)))...(((((......((((.(((((..((.((((.........)))).))..))))).)))-------)....)))))................ ( -27.40) >DroSim_CAF1 128600 112 - 1 UUUGGCGCAUUCUUCUAAUUGCCCCACUGUGGCAGAAACAGAAUCUGCAAUCAGCGCUGCAUCACUUACAGCACUUUUUGCAAAUU-------CCACUUGCCACUUUUUGGCAUUUCGU ...................((((.....(((((((.....((((.(((((..((.((((.........)))).))..))))).)))-------)...))))))).....))))...... ( -31.30) >DroEre_CAF1 129876 111 - 1 UUUGGCGCAUUCUUCUAAUUGCCCAACUGUGGCAGAAACAGAAUUCGCAAUCAGCGCCGCAUCACUUACAGCACUUUUUGCAAUU--------CCACUUGCCACUUUUUGGCAUCUCGU ...................((((.((..(((((((.....(((((.((((..((.((.(.........).)).))..))))))))--------)...)))))))..)).))))...... ( -26.20) >DroYak_CAF1 130683 107 - 1 UUUGGCGCAUUCUUCUAAUUGCCCGGCUGUGGCAGAAACAGAAUCUGCAAUCAGCGC------------AGCACUUUUUGCAAAUGCCACUUGCCACUUGCCACUUUUUGGCAUUUCGU ...((.(((..........)))))(((.((((((....(((((.((((.......))------------))....)))))....))))))..)))...(((((.....)))))...... ( -33.90) >consensus UUUGGCGCAUUCUUCUAAUUGCCCCACUGUGGCAGAAACAGAAUCUGCAAUCAGCGCUGCAUCACUUACAGCACUUUUUGCAAAUU_______CCACUUGCCACUUUUUGGCAUUUCGU ..((((..(((((.....((((((....).)))))....)))))..((((..((.((((.........)))).))..))))..................))))................ (-21.17 = -21.82 + 0.65)


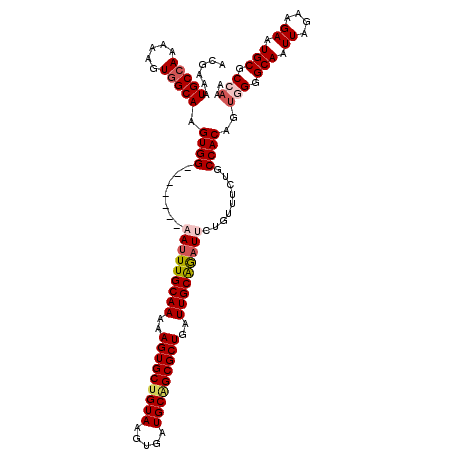
| Location | 18,765,258 – 18,765,366 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -35.46 |
| Consensus MFE | -28.93 |
| Energy contribution | -30.12 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18765258 108 + 20766785 AAAAAGUGCUGUAAGUGAUGCAGCGCUGAUUGCAGAUUCUGUUUCUGCCACAGUGGGGCAAUUAGAAGAAUGCGCCAAAUGGGCUCCAAAGCCCGGUUGAGCGCGGCC ....(((((((((.....)))))))))....((..(((((.....((((.......))))......)))))(((((((.((((((....)))))).))).)))).)). ( -41.40) >DroSec_CAF1 127777 108 + 1 AAAAAGUGCUGUAAGUGAUGCAGCGCUGAUUGCAGAUUCUGUUUAUGCCACAGUGGGGCAAUUAGAAGAAUGCGCCAAAUGGGCUCCAAAGCUCGACUGAGCCCGGCC ....(((((((((.....)))))))))....(((..(((((....((((.......))))..)))))...)))(((....((((((............))))))))). ( -37.80) >DroSim_CAF1 128633 108 + 1 AAAAAGUGCUGUAAGUGAUGCAGCGCUGAUUGCAGAUUCUGUUUCUGCCACAGUGGGGCAAUUAGAAGAAUGCGCCAAAUGGGCUCCGAAGCUCGGCUGAGCACGGCC ....(((((((((.....)))))))))....(((((.......))))).....(((.(((.((....)).))).)))...(((((....)))))(((((....))))) ( -37.30) >DroEre_CAF1 129908 95 + 1 AAAAAGUGCUGUAAGUGAUGCGGCGCUGAUUGCGAAUUCUGUUUCUGCCACAGUUGGGCAAUUAGAAGAAUGCGCCAAAUGGG-------------UUGAGAUCGGCC ....(((((((((.....)))))))))....(((.(((((.....((((.(....)))))......))))).)))......((-------------(((....))))) ( -28.90) >DroYak_CAF1 130723 96 + 1 AAAAAGUGCU------------GCGCUGAUUGCAGAUUCUGUUUCUGCCACAGCCGGGCAAUUAGAAGAAUGCGCCAAAUGGGCUCCGUAACUCGGUGGAGCUCGGCC ........((------------(.((((...(((((.......)))))..)))))))(((.((....)).)))(((....((((((((........))))))))))). ( -31.90) >consensus AAAAAGUGCUGUAAGUGAUGCAGCGCUGAUUGCAGAUUCUGUUUCUGCCACAGUGGGGCAAUUAGAAGAAUGCGCCAAAUGGGCUCCAAAGCUCGGUUGAGCUCGGCC ....(((((((((.....)))))))))....(((..(((((....((((.......))))..)))))...)))(((....((((((............))))))))). (-28.93 = -30.12 + 1.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:51 2006