
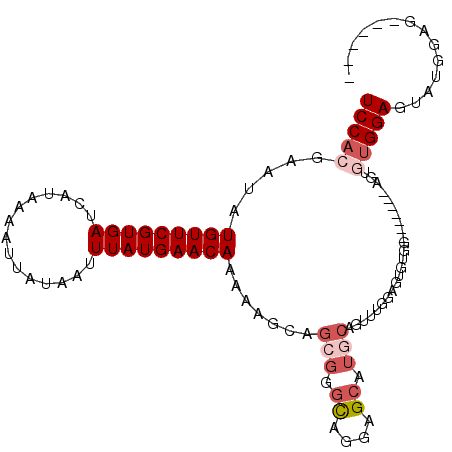
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,747,742 – 18,747,845 |
| Length | 103 |
| Max. P | 0.999011 |

| Location | 18,747,742 – 18,747,845 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 77.60 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -11.96 |
| Energy contribution | -13.40 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
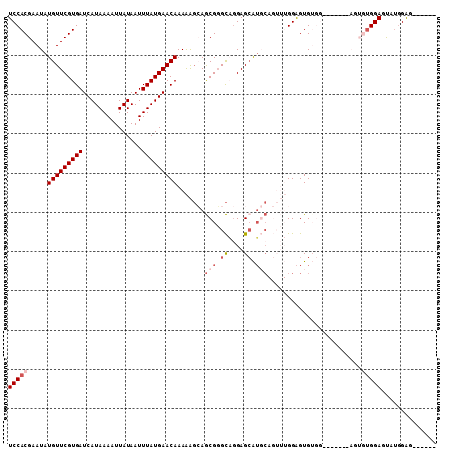
>2R_DroMel_CAF1 18747742 103 + 20766785 UCCACGAAUAUGUUCGUGAUCAUAAAAUUAUAAUUUAUGAACAAAAAGCAGCGGGCAGGAGCAUGCAGUUUGGAGUGUGGAGUCUGGAGUGUGGAGUAUGGAG------ (((((...(((((((.((.(((((((.......))))))).))....((.....))..))))))).(.((..((.(....).))..)).))))))........------ ( -23.80) >DroSec_CAF1 110495 89 + 1 UCCACGAAUAUGUUCGUGAUCAUAAAAUUAUAAUUUAUGAACAAAAAGCAGCGGGCAGGAGCAUGCAGUUUGGAGUG--------------UGGAGUAUGGAG------ ((((((....(((((((((...............)))))))))..((((.(((.((....)).))).))))....))--------------))))........------ ( -21.46) >DroSim_CAF1 111270 96 + 1 UCCACGAAUAUGUUCGUGAUCAUAAAAUUAUAAUUUAUGAACAAAAAGCAGCGGGCAGGAGCAUGCAGUUUGGAGUGUGG-------AGUGUGGAGUAUGGAG------ ((((((..((..(((....(((((((.......))))))).((((..(((((........)).)))..)))))))..)).-------..))))))........------ ( -25.80) >DroMoj_CAF1 171624 94 + 1 UCCACGAAUAUGUUCGUGAUCAUAAAAUUAUAAUUUAUGAACAAAGGCGAACGCGU-GGAGCAU-------GGAGUGUGG-------AGUGUGGAGCGUGGAGCGGCCC ((((((..(((((((.((.(((((((.......))))))).))...((....))..-.))))))-------)...)))))-------)....((.((.....))..)). ( -26.80) >DroAna_CAF1 107089 80 + 1 UCCACGAAUAUGUUCGUGAUCAUAAAAUUAUAAUUUAUGAACAAAAAGAAGCGGGCCAGAGGGGCC-----GGGGUG---------------GGACU---GGG------ (((((.....(((((((((...............)))))))))........(.((((.....))))-----.).)))---------------))...---...------ ( -22.66) >consensus UCCACGAAUAUGUUCGUGAUCAUAAAAUUAUAAUUUAUGAACAAAAAGCAGCGGGCAGGAGCAUGCAGUUUGGAGUGUGG_______AGUGUGGAGUAUGGAG______ (((((.....(((((((((...............))))))))).......(((.((....)).)))........................))))).............. (-11.96 = -13.40 + 1.44)

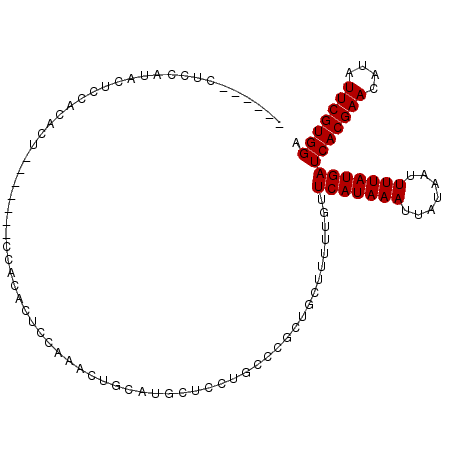
| Location | 18,747,742 – 18,747,845 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 77.60 |
| Mean single sequence MFE | -18.86 |
| Consensus MFE | -10.80 |
| Energy contribution | -10.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.57 |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.999011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
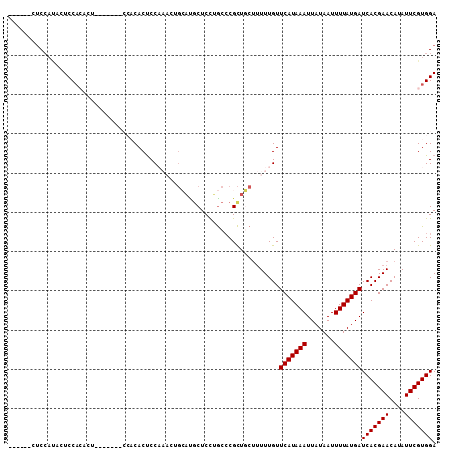
>2R_DroMel_CAF1 18747742 103 - 20766785 ------CUCCAUACUCCACACUCCAGACUCCACACUCCAAACUGCAUGCUCCUGCCCGCUGCUUUUUGUUCAUAAAUUAUAAUUUUAUGAUCACGAACAUAUUCGUGGA ------......................(((((....((((..(((.((........)))))..)))).(((((((.......)))))))..............))))) ( -18.10) >DroSec_CAF1 110495 89 - 1 ------CUCCAUACUCCA--------------CACUCCAAACUGCAUGCUCCUGCCCGCUGCUUUUUGUUCAUAAAUUAUAAUUUUAUGAUCACGAACAUAUUCGUGGA ------........((((--------------(....((((..(((.((........)))))..)))).(((((((.......)))))))..............))))) ( -18.10) >DroSim_CAF1 111270 96 - 1 ------CUCCAUACUCCACACU-------CCACACUCCAAACUGCAUGCUCCUGCCCGCUGCUUUUUGUUCAUAAAUUAUAAUUUUAUGAUCACGAACAUAUUCGUGGA ------...............(-------((((....((((..(((.((........)))))..)))).(((((((.......)))))))..............))))) ( -18.10) >DroMoj_CAF1 171624 94 - 1 GGGCCGCUCCACGCUCCACACU-------CCACACUCC-------AUGCUCC-ACGCGUUCGCCUUUGUUCAUAAAUUAUAAUUUUAUGAUCACGAACAUAUUCGUGGA ((..((.....))..))....(-------((((.....-------..((...-..))(((((....((.(((((((.......))))))).)))))))......))))) ( -22.10) >DroAna_CAF1 107089 80 - 1 ------CCC---AGUCC---------------CACCCC-----GGCCCCUCUGGCCCGCUUCUUUUUGUUCAUAAAUUAUAAUUUUAUGAUCACGAACAUAUUCGUGGA ------...---....(---------------(((...-----((((.....))))........((((((((((((.......)))))))..))))).......)))). ( -17.90) >consensus ______CUCCAUACUCCACACU_______CCACACUCCAAACUGCAUGCUCCUGCCCGCUGCUUUUUGUUCAUAAAUUAUAAUUUUAUGAUCACGAACAUAUUCGUGGA .....................................................................(((((((.......)))))))(((((((....))))))). (-10.80 = -10.80 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:43 2006