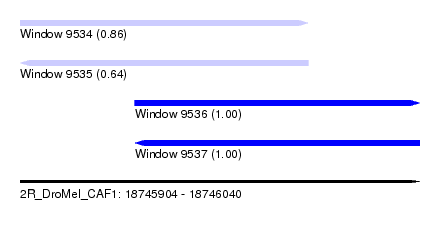
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,745,904 – 18,746,040 |
| Length | 136 |
| Max. P | 0.998881 |

| Location | 18,745,904 – 18,746,002 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 88.31 |
| Mean single sequence MFE | -30.34 |
| Consensus MFE | -23.46 |
| Energy contribution | -23.86 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
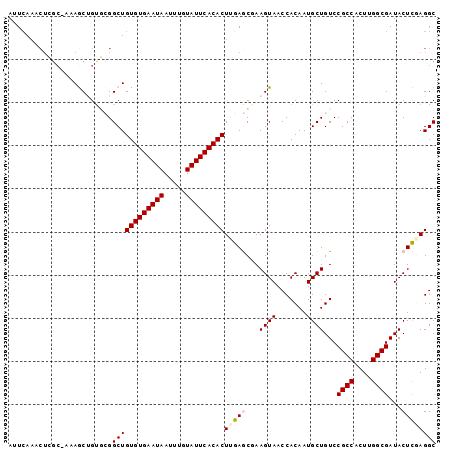
>2R_DroMel_CAF1 18745904 98 + 20766785 GCCUCGAGUAUCGCCAAGUGGCGGACAGCAUUGUGGUUACUUCGCUCAAGUGUGAAUACAAAUUAUUCACACAGCCGCACAGCUUU-GCGAGUUUGAAU ...((((...((((.((((.((((.(......((((.....))))....(((((((((.....))))))))).)))))...)))).-))))..)))).. ( -33.20) >DroSec_CAF1 108634 98 + 1 GCCUGGAGUAUCGCCAAGUGGCGGACAGCAUUGUGGUUACUUGGCUCAAGUGUGAAUACAAAUUAUUCACACAGCCGCACAGCUUU-GCGAGUUUGAAU ............((((((((((.(.(......)).))))))))))(((((((((((((.....))))))))).((((((......)-))).)))))).. ( -32.70) >DroSim_CAF1 109418 98 + 1 GCCUGGAGUAUCGCCAAGUGGCGGACAGCAUUGUGGUUACUUGGCUCAAGUGUGAAUACAAAUUAUUCACACAGCCGCACAGCUUU-GCGAGUUUGAAU ............((((((((((.(.(......)).))))))))))(((((((((((((.....))))))))).((((((......)-))).)))))).. ( -32.70) >DroEre_CAF1 111099 89 + 1 GCCUCAAGUAUCGCCGAGUGGCGGACAGCAUUGUGGUUACUUCGUUUGAGUGUGAAUACAAAUUAUUCACACAGCCCAACCGUU----------UGAAU ...((((((.(((((....)))))))....(((.((((...((....))(((((((((.....))))))))))))))))....)----------))).. ( -23.70) >DroYak_CAF1 111342 99 + 1 GCCUCAAGUAUCGCCAAGUGGCGGACAGCAUUGUGGUUACUUCGUUUGAGUGUGAAUACAAAUUAUUCACACAGCCCAGCCGUUUUGGCUAGUUUCAAU (((.(((((.(((((....)))))...)).))).)))........(((((((((((((.....))))))))).....((((.....))))....)))). ( -29.40) >consensus GCCUCGAGUAUCGCCAAGUGGCGGACAGCAUUGUGGUUACUUCGCUCAAGUGUGAAUACAAAUUAUUCACACAGCCGCACAGCUUU_GCGAGUUUGAAU .......((.(((((....)))))))(((..(((((((...........(((((((((.....))))))))))))))))..)))............... (-23.46 = -23.86 + 0.40)

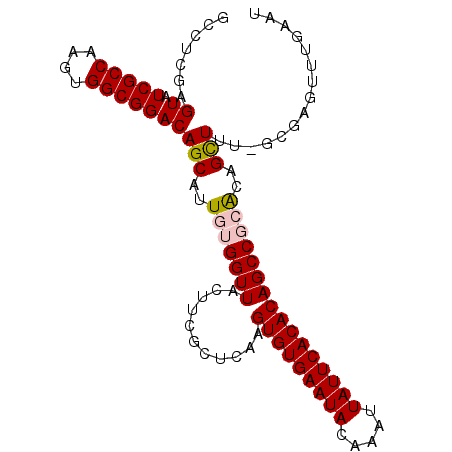

| Location | 18,745,904 – 18,746,002 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 88.31 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -20.04 |
| Energy contribution | -20.28 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18745904 98 - 20766785 AUUCAAACUCGC-AAAGCUGUGCGGCUGUGUGAAUAAUUUGUAUUCACACUUGAGCGAAGUAACCACAAUGCUGUCCGCCACUUGGCGAUACUCGAGGC .......(((((-..((((....))))(((((((((.....)))))))))....))((.(((...(((....))).((((....)))).)))))))).. ( -28.30) >DroSec_CAF1 108634 98 - 1 AUUCAAACUCGC-AAAGCUGUGCGGCUGUGUGAAUAAUUUGUAUUCACACUUGAGCCAAGUAACCACAAUGCUGUCCGCCACUUGGCGAUACUCCAGGC ........((((-.(((.((.(((((((((((((((.....)))))))))...)))).((((.......))))....))))))).)))).......... ( -27.70) >DroSim_CAF1 109418 98 - 1 AUUCAAACUCGC-AAAGCUGUGCGGCUGUGUGAAUAAUUUGUAUUCACACUUGAGCCAAGUAACCACAAUGCUGUCCGCCACUUGGCGAUACUCCAGGC ........((((-.(((.((.(((((((((((((((.....)))))))))...)))).((((.......))))....))))))).)))).......... ( -27.70) >DroEre_CAF1 111099 89 - 1 AUUCA----------AACGGUUGGGCUGUGUGAAUAAUUUGUAUUCACACUCAAACGAAGUAACCACAAUGCUGUCCGCCACUCGGCGAUACUUGAGGC .((((----------(((((((((((((((((((((.....)))))))))((....)))))..)))....))))).((((....))))....))))).. ( -23.80) >DroYak_CAF1 111342 99 - 1 AUUGAAACUAGCCAAAACGGCUGGGCUGUGUGAAUAAUUUGUAUUCACACUCAAACGAAGUAACCACAAUGCUGUCCGCCACUUGGCGAUACUUGAGGC .((((..((((((.....))))))...(((((((((.....))))))))))))).........((.(((.(.((((.(((....))))))))))).)). ( -31.40) >consensus AUUCAAACUCGC_AAAGCUGUGCGGCUGUGUGAAUAAUUUGUAUUCACACUUGAGCGAAGUAACCACAAUGCUGUCCGCCACUUGGCGAUACUCGAGGC ........................((((((((((((.....)))))))))(((((...((((.......))))...((((....))))...)))))))) (-20.04 = -20.28 + 0.24)

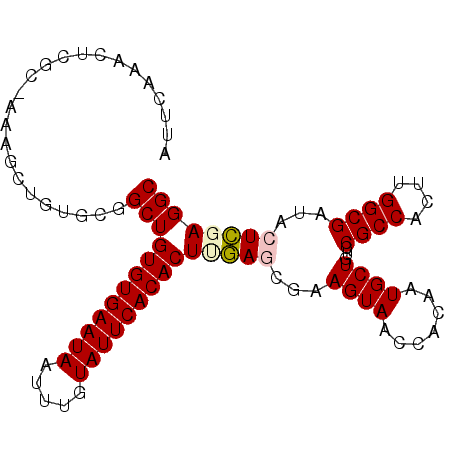
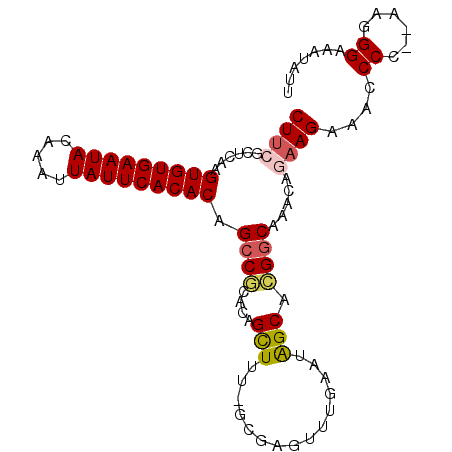
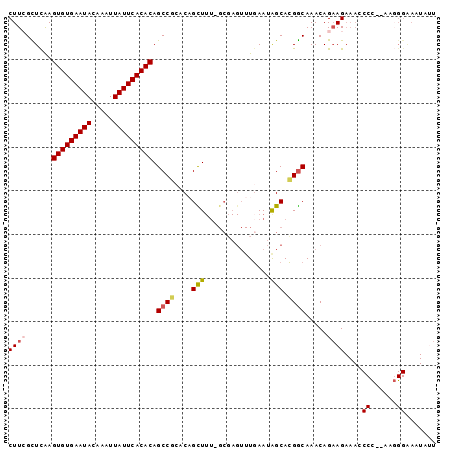
| Location | 18,745,943 – 18,746,040 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 77.29 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -15.68 |
| Energy contribution | -16.06 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.61 |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18745943 97 + 20766785 CUUCGCUCAAGUGUGAAUACAAAUUAUUCACACAGCCGCACAGCUUU-GCGAGUUUGAAUAGCACGGCAAACAGAAGAAACCCCCAAAGGGAAAUAAU ((((......(((((((((.....))))))))).((((((......)-))..(((.....)))..))).....))))...(((.....)))....... ( -26.70) >DroSec_CAF1 108673 95 + 1 CUUGGCUCAAGUGUGAAUACAAAUUAUUCACACAGCCGCACAGCUUU-GCGAGUUUGAAUAGCACGGCAAACAGAAGAAAGCCC--AAGGGAAAUAUU (((((((...(((((((((.....))))))))).((((((......)-))..(((.....)))..)))...........)).))--)))......... ( -26.00) >DroSim_CAF1 109457 95 + 1 CUUGGCUCAAGUGUGAAUACAAAUUAUUCACACAGCCGCACAGCUUU-GCGAGUUUGAAUAGCACGGCAAACAGAAGAAAGCCC--AAGGGAAAUAUU (((((((...(((((((((.....))))))))).((((((......)-))..(((.....)))..)))...........)).))--)))......... ( -26.00) >DroEre_CAF1 111138 84 + 1 CUUCGUUUGAGUGUGAAUACAAAUUAUUCACACAGCCCAACCGUU----------UGAAUGGCACGGCAAACAGAAGAAACCCU--A--GGGAAUAUU ((((((((..(((((((((.....))))))))).(((...((((.----------...))))...))))))).))))...((..--.--.))...... ( -25.40) >DroYak_CAF1 111381 77 + 1 CUUCGUUUGAGUGUGAAUACAAAUUAUUCACACAGCCCAGCCGUUUUGGCUAGUUUCAAUGGCAGGACAAACAGUAG--------------------- ....(((((.(((((((((.....)))))))))...((.((((((..((....))..)))))).)).))))).....--------------------- ( -24.40) >consensus CUUCGCUCAAGUGUGAAUACAAAUUAUUCACACAGCCGCACAGCUUU_GCGAGUUUGAAUAGCACGGCAAACAGAAGAAACCCC__AAGGGAAAUAUU ((((......(((((((((.....))))))))).((((....(((...............))).)))).....))))....((......))....... (-15.68 = -16.06 + 0.38)

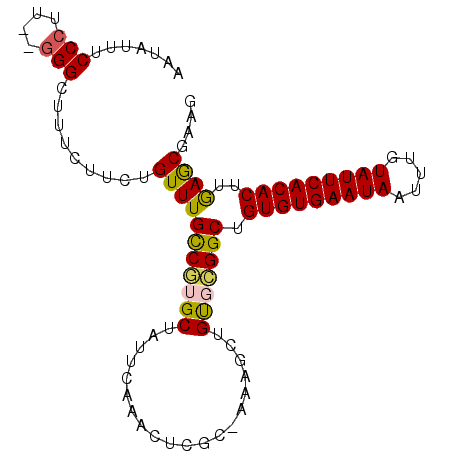
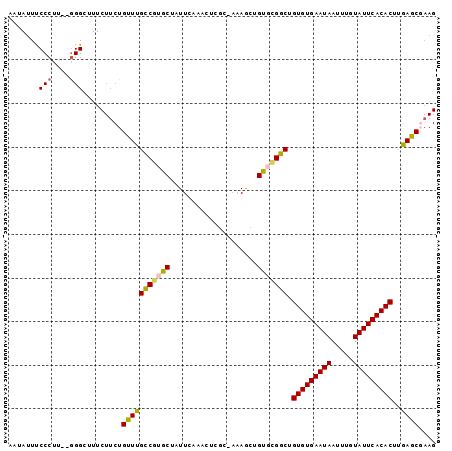
| Location | 18,745,943 – 18,746,040 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 77.29 |
| Mean single sequence MFE | -26.11 |
| Consensus MFE | -20.72 |
| Energy contribution | -20.63 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18745943 97 - 20766785 AUUAUUUCCCUUUGGGGGUUUCUUCUGUUUGCCGUGCUAUUCAAACUCGC-AAAGCUGUGCGGCUGUGUGAAUAAUUUGUAUUCACACUUGAGCGAAG ......((((....))))...((((.((((((((..(.............-......)..)))).(((((((((.....)))))))))..)))))))) ( -29.11) >DroSec_CAF1 108673 95 - 1 AAUAUUUCCCUU--GGGCUUUCUUCUGUUUGCCGUGCUAUUCAAACUCGC-AAAGCUGUGCGGCUGUGUGAAUAAUUUGUAUUCACACUUGAGCCAAG ............--.(((((..........((((..(.............-......)..)))).(((((((((.....)))))))))..)))))... ( -27.21) >DroSim_CAF1 109457 95 - 1 AAUAUUUCCCUU--GGGCUUUCUUCUGUUUGCCGUGCUAUUCAAACUCGC-AAAGCUGUGCGGCUGUGUGAAUAAUUUGUAUUCACACUUGAGCCAAG ............--.(((((..........((((..(.............-......)..)))).(((((((((.....)))))))))..)))))... ( -27.21) >DroEre_CAF1 111138 84 - 1 AAUAUUCCC--U--AGGGUUUCUUCUGUUUGCCGUGCCAUUCA----------AACGGUUGGGCUGUGUGAAUAAUUUGUAUUCACACUCAAACGAAG ......((.--.--..))...((((.(((((((..(((.....----------...)))..))).(((((((((.....)))))))))..)))))))) ( -23.20) >DroYak_CAF1 111381 77 - 1 ---------------------CUACUGUUUGUCCUGCCAUUGAAACUAGCCAAAACGGCUGGGCUGUGUGAAUAAUUUGUAUUCACACUCAAACGAAG ---------------------....((((((..............((((((.....))))))...(((((((((.....))))))))).))))))... ( -23.80) >consensus AAUAUUUCCCUU__GGGCUUUCUUCUGUUUGCCGUGCUAUUCAAACUCGC_AAAGCUGUGCGGCUGUGUGAAUAAUUUGUAUUCACACUUGAGCGAAG .......(((....))).........(((((((((((....................))))))).(((((((((.....)))))))))..)))).... (-20.72 = -20.63 + -0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:41 2006