| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,743,927 – 18,744,020 |
| Length | 93 |
| Max. P | 0.998777 |

| Location | 18,743,927 – 18,744,020 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.68 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -19.50 |
| Energy contribution | -19.38 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18743927 93 + 20766785 CGUCGCCGCU-----------GCUGCCGGUAAAUGUCUUUAUAGACAUUAUUCCCGC--AGUCGCCUCGUCUUGGCAAAGCUUUAAAUUUAUGGCAACGCAUUUGC (((.((((..-----------(((((.((..(((((((....)))))))...)).))--))).(((.......)))...............)))).)))....... ( -29.00) >DroEre_CAF1 109153 92 + 1 CGCCGUCGCC-----------GCUGCCGGUAAAUGUCUUUAUAGGCAUUAUUCCCGCACAGUCGCC---UCCUGGCAAAGCUUUAAAUUUAUGGCAACGCAUUUGC .((.((.(((-----------(.(((.((..(((((((....)))))))...)).))))....(((---....)))................))).))))...... ( -25.90) >DroYak_CAF1 109322 102 + 1 CGCCGUCGUUGCCGCAUUGGAGCUGCCGGUAAAUGUCUUUAUAGACAUUAUUCCCGC--CGUCGCCU--UCCUGGCAAAGCUUUAAAUUUAUGGCAACGCAUUUGC ....(.((((((((..((((((((((.((..(((((((....)))))))...)).))--....(((.--....)))..)))))))).....)))))))))...... ( -35.30) >DroAna_CAF1 103492 75 + 1 ---------------------GUUGCCGGUAAAUGUCUUCAUAGACAU---UCCCGG--AGUCGCCG-----CGGCAAAGCUUUAAAUUUAUGGCAACGCAUUUGC ---------------------((((((((..(((((((....))))))---).))((--(((.((..-----..))...)))))........))))))........ ( -22.90) >consensus CGCCGUCGCU___________GCUGCCGGUAAAUGUCUUUAUAGACAUUAUUCCCGC__AGUCGCCU__UCCUGGCAAAGCUUUAAAUUUAUGGCAACGCAUUUGC .....................((((((((..(((((((....)))))))...)).........(((.......)))................))))..))...... (-19.50 = -19.38 + -0.12)

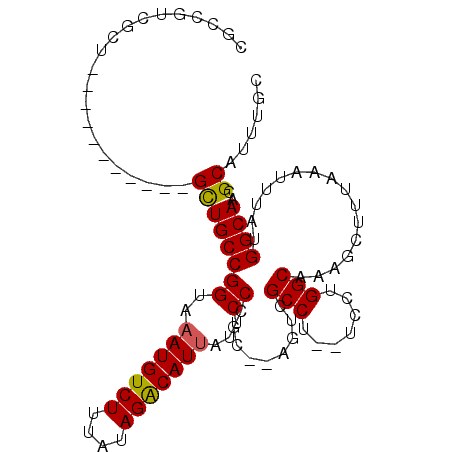
| Location | 18,743,927 – 18,744,020 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.68 |
| Mean single sequence MFE | -31.25 |
| Consensus MFE | -23.56 |
| Energy contribution | -23.88 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.75 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18743927 93 - 20766785 GCAAAUGCGUUGCCAUAAAUUUAAAGCUUUGCCAAGACGAGGCGACU--GCGGGAAUAAUGUCUAUAAAGACAUUUACCGGCAGC-----------AGCGGCGACG ((....))((((((...........((((((......))))))(.((--((.((...(((((((....)))))))..)).)))))-----------...)))))). ( -36.00) >DroEre_CAF1 109153 92 - 1 GCAAAUGCGUUGCCAUAAAUUUAAAGCUUUGCCAGGA---GGCGACUGUGCGGGAAUAAUGCCUAUAAAGACAUUUACCGGCAGC-----------GGCGACGGCG .......(((((((..............(((((....---)))))((((.(((....((((.((....)).))))..))))))).-----------)))))))... ( -28.20) >DroYak_CAF1 109322 102 - 1 GCAAAUGCGUUGCCAUAAAUUUAAAGCUUUGCCAGGA--AGGCGACG--GCGGGAAUAAUGUCUAUAAAGACAUUUACCGGCAGCUCCAAUGCGGCAACGACGGCG ((.....(((((((..............(((((....--.))))).(--.(((....(((((((....)))))))..))).).((......)))))))))...)). ( -36.30) >DroAna_CAF1 103492 75 - 1 GCAAAUGCGUUGCCAUAAAUUUAAAGCUUUGCCG-----CGGCGACU--CCGGGA---AUGUCUAUGAAGACAUUUACCGGCAAC--------------------- ((....))((((((..............((((..-----..))))..--..((((---((((((....)))))))).))))))))--------------------- ( -24.50) >consensus GCAAAUGCGUUGCCAUAAAUUUAAAGCUUUGCCAGGA__AGGCGACU__GCGGGAAUAAUGUCUAUAAAGACAUUUACCGGCAGC___________AGCGACGGCG ((....))((((((..............(((((.......)))))......((....(((((((....)))))))..))))))))..................... (-23.56 = -23.88 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:37 2006