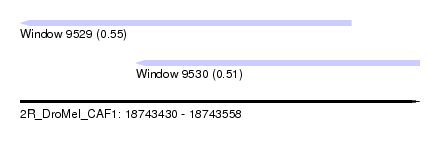
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,743,430 – 18,743,558 |
| Length | 128 |
| Max. P | 0.549548 |

| Location | 18,743,430 – 18,743,536 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.45 |
| Mean single sequence MFE | -40.12 |
| Consensus MFE | -22.05 |
| Energy contribution | -22.83 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
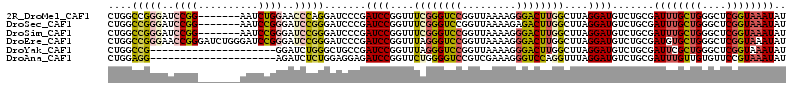
>2R_DroMel_CAF1 18743430 106 - 20766785 CUGGCCGGGAUCCGG-------AAUCUGGAACCCAGGAUCCCGAUCCGGUUUCGGGUCCGGUUAAAAGGGACUUGGCUUAGGAUGUCUGCGAUUUGCUGGGCUCGGUAAAUAU .((((((((.(((((-------...))))).))).(((.(((((.......)))))))))))))......(((.(((((((.(.(((...))).).))))))).)))...... ( -40.50) >DroSec_CAF1 106159 106 - 1 CUGGCCGGGAUCCGG-------AAUCCGGGAUCCGGGAUCCCGAUCCGGUUUCGGGUCCGGUUAAAAGAGACUUGGCUUAGGAUGUCUGCGAUUUGCUGGGCUCGGUAAAUAU .(((((((.((((((-------((.((((.(((.((....))))))))))))))))))))))))......(((.(((((((.(.(((...))).).))))))).)))...... ( -44.80) >DroSim_CAF1 106938 106 - 1 CUGGCCGGGAUCCGG-------AAUCCGGGAUCCGGGAUCCCGAUCCGGUUUCGGGUCCGGUUAAAAGGGACUUGGCUUAGGAUGUCUGCGAUUUGCUGGGCUCGGUAAAUAU .(((((((.((((((-------((.((((.(((.((....))))))))))))))))))))))))......(((.(((((((.(.(((...))).).))))))).)))...... ( -44.80) >DroEre_CAF1 108650 113 - 1 CUGGCCGGGAACCGGGAUCUGGGAUCCGGGAUCCGGGAUCCCGAUCCGGUUUAGGGUCCGGUUAAAAGGGACUUGGCUUAGGAUGUCUGCGAUGUGCUGGGCUCGGUAAAUAU ((((((..(((((((.(((.(((((((.(....).)))))))))))))))))..)).)))).........(((.(((((((.(((((...))))).))))))).)))...... ( -52.80) >DroYak_CAF1 108845 92 - 1 CUGGCCG---------------------GGAUCUGGGCUGCCGAUCCGGUUUAGGGUCCGGUUAAAAGGGACUUGGCUUAGGAUGUCUGCGAUUCGCUGGGCUCGGUAAAUAU .((((((---------------------((..((((((((......))))))))..))))))))......(((.(((((((((.(((...))))).))))))).)))...... ( -31.60) >DroAna_CAF1 103020 92 - 1 CUGGAGG---------------------AGAUCUCUGGAGGAGAUCCGGUUCUGGGGUCCGUCGAAAGGGUCCAGGUUUAGGAUGUCUGCGAUUUGUUGUGUUCCGUAAAUAU ..((((.---------------------....))))((((((.((((....((.((..((.......))..)).))....)))).)))((((....))))..)))........ ( -26.20) >consensus CUGGCCGGGAUCCGG_______AAUCCGGGAUCCGGGAUCCCGAUCCGGUUUCGGGUCCGGUUAAAAGGGACUUGGCUUAGGAUGUCUGCGAUUUGCUGGGCUCGGUAAAUAU ....(((((..((((..........))))..))))).......((((....((((((((.........))))))))....)))).......((((((((....)))))))).. (-22.05 = -22.83 + 0.78)
| Location | 18,743,467 – 18,743,558 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 85.18 |
| Mean single sequence MFE | -34.94 |
| Consensus MFE | -21.29 |
| Energy contribution | -23.20 |
| Covariance contribution | 1.91 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
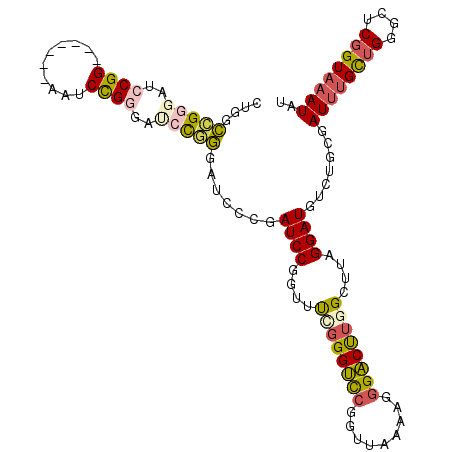
>2R_DroMel_CAF1 18743467 91 - 20766785 CACCUGUUACACUCAUCUUUCUCUGGCCGGGAUCCGG-------AAUCUGGAACCCAGGAUCCCGAUCCGGUUUCGGGUCCGGUUAAAAGGGACUUGG .............((..(..((.((((((((.(((((-------...))))).))).(((.(((((.......)))))))))))))..))..)..)). ( -32.20) >DroSec_CAF1 106196 91 - 1 CACCUGUUACACUCAUCUUUUUCUGGCCGGGAUCCGG-------AAUCCGGGAUCCGGGAUCCCGAUCCGGUUUCGGGUCCGGUUAAAAGAGACUUGG ...............(((((((.(((((((.((((((-------((.((((.(((.((....)))))))))))))))))))))))))))))))..... ( -38.20) >DroSim_CAF1 106975 91 - 1 CACCUGUUACACUCAUCUUUUUCUGGCCGGGAUCCGG-------AAUCCGGGAUCCGGGAUCCCGAUCCGGUUUCGGGUCCGGUUAAAAGGGACUUGG ...............(((((((.(((((((.((((((-------((.((((.(((.((....)))))))))))))))))))))))))))))))..... ( -37.40) >DroEre_CAF1 108687 97 - 1 CACCUGUUACACUCAGCUU-UUCUGGCCGGGAACCGGGAUCUGGGAUCCGGGAUCCGGGAUCCCGAUCCGGUUUAGGGUCCGGUUAAAAGGGACUUGG ...........((.(((((-((.((((((((((((((.(((.(((((((.(....).))))))))))))))))).....))))))).))))).)).)) ( -42.10) >DroYak_CAF1 108882 77 - 1 CACCUGUUACACUCAUCUUUUUCUGGCCG---------------------GGAUCUGGGCUGCCGAUCCGGUUUAGGGUCCGGUUAAAAGGGACUUGG ...............(((((((.((((((---------------------((..((((((((......))))))))..)))))))))))))))..... ( -24.80) >consensus CACCUGUUACACUCAUCUUUUUCUGGCCGGGAUCCGG_______AAUCCGGGAUCCGGGAUCCCGAUCCGGUUUCGGGUCCGGUUAAAAGGGACUUGG ...............(((((((.((((((((.(((((..........))))).))).(((.(((((.......))))))))))))))))))))..... (-21.29 = -23.20 + 1.91)

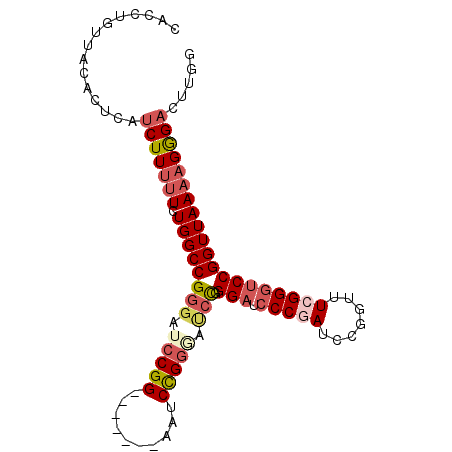
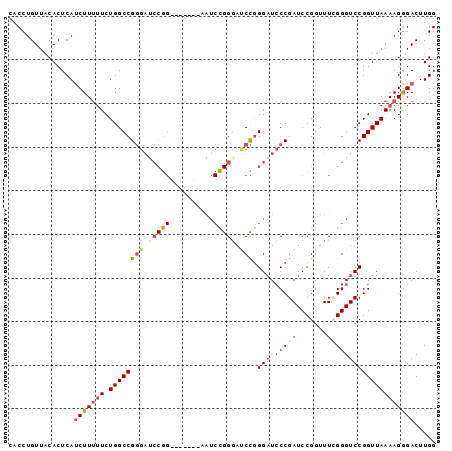
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:35 2006