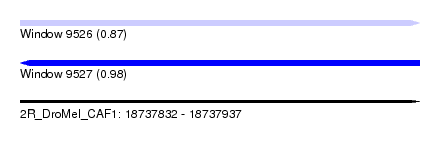
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,737,832 – 18,737,937 |
| Length | 105 |
| Max. P | 0.976672 |

| Location | 18,737,832 – 18,737,937 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.00 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -13.20 |
| Energy contribution | -13.87 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.874100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
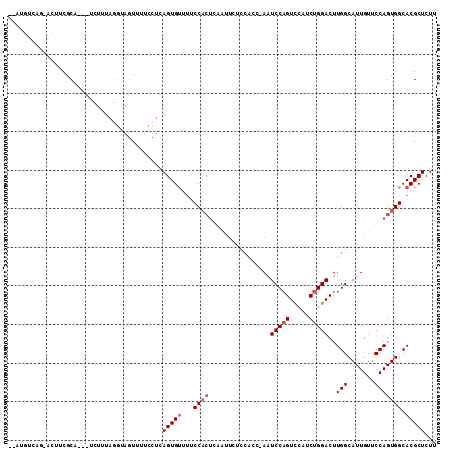
>2R_DroMel_CAF1 18737832 105 + 20766785 --AUGUCAGUAUGUCGCA---UCUUUAGUAGGUUUUCCUCAGUGUUUUCCACUCAAUUCUCCACC-AAUCCAGUCCAUCUGGACUUGGCAUUGUUCCAGUGGCACGCUCUU --.....((..((((((.---.........((........((((.....)))).......))..(-((((((((((....)))).))).)))).....))))))..))... ( -25.56) >DroSec_CAF1 100648 105 + 1 --AUGUCAGUACUUCGCA---UCUGUAGGUAGUUUUCCUCAGUGUUUUCCACUCAAUUCUCCAAC-AAUCCAGUCCAUCUGGACUUGGCAUUGUUCCAGUGGCACGCUCUU --.(((((.(.....(((---.(((.(((.......))))))))).................(((-((((((((((....)))).))).))))))..).)))))....... ( -25.00) >DroSim_CAF1 101439 105 + 1 --AUGUGAGUACUUCGCA---UCUGUAGGUAGUUUUCCUUAGUGUUUUCCACUCAAUUCUCCAAC-AAUCCAGUCCAUCUGGACUUGGCAUUGUUCCAGUGGCACGCUCUU --....((((.....(((---.(((.(((.......)))))))))...(((((.........(((-((((((((((....)))).))).))))))..)))))...)))).. ( -25.40) >DroEre_CAF1 103279 93 + 1 --------------CACU----UUUUAAGUGGUUUCCCUCAGUGUUUUCCACUCAACUAUCCACUAAAUCCAGUUGAUCUGGACUUGGCAUUGUUCCAGUGGCACGCUCUA --------------((((----.....)))).........(((((...(((((.(((......((((.(((((.....))))).))))....)))..))))).)))))... ( -21.20) >DroYak_CAF1 103106 94 + 1 UAAUGUCAA-GCUUCACC----UUUUAGGUAGUUUUCCUCAGUGUUUUCC------------ACUAAAUCCAGUCGAUCUGGACUUGGCAUUGUGCCAGUGGCACGCUCUU .((((((((-(....(((----.....)))..........((((.....)------------)))...(((((.....))))))))))))))(((((...)))))...... ( -27.00) >DroAna_CAF1 97676 97 + 1 --A---------CUCGCAGGGUGUCUAUGGGAUUUCCCCAAGUGCACCCCACUCUGUUGACC---GAAUCCCGUCCUUCUGGACUUGGCAUUGUUCCAGUGGCACGCUCUC --.---------...((.((((((((.((((.....)))))).))))))((((....((.((---((.....((((....)))))))))).......))))))........ ( -29.10) >consensus __AUGUCAG_ACUUCGCA___UCUUUAGGUAGUUUUCCUCAGUGUUUUCCACUCAAUUCUCCACC_AAUCCAGUCCAUCUGGACUUGGCAUUGUUCCAGUGGCACGCUCUU ........................................(((((...((((................(((((.....)))))..(((.......))))))).)))))... (-13.20 = -13.87 + 0.67)

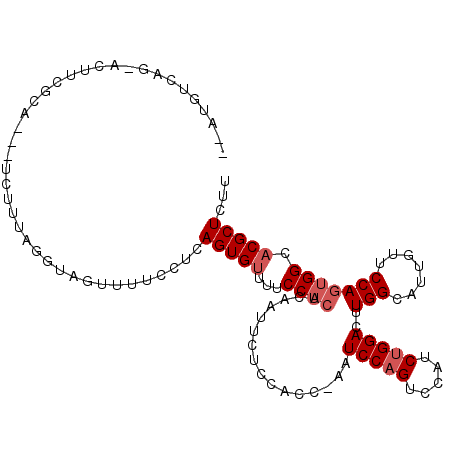
| Location | 18,737,832 – 18,737,937 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 74.00 |
| Mean single sequence MFE | -28.97 |
| Consensus MFE | -15.53 |
| Energy contribution | -15.45 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
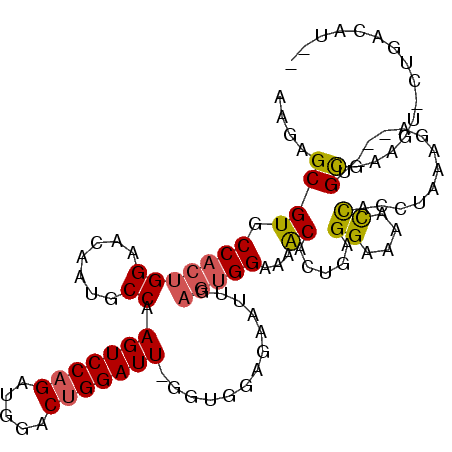
>2R_DroMel_CAF1 18737832 105 - 20766785 AAGAGCGUGCCACUGGAACAAUGCCAAGUCCAGAUGGACUGGAUU-GGUGGAGAAUUGAGUGGAAAACACUGAGGAAAACCUACUAAAGA---UGCGACAUACUGACAU-- ....((((.(((((.((....((((((.(((((.....)))))))-)))).....)).))))).........(((....))).......)---))).............-- ( -26.30) >DroSec_CAF1 100648 105 - 1 AAGAGCGUGCCACUGGAACAAUGCCAAGUCCAGAUGGACUGGAUU-GUUGGAGAAUUGAGUGGAAAACACUGAGGAAAACUACCUACAGA---UGCGAAGUACUGACAU-- ....((((.(((((.(((((((.(((.((((....))))))))))-)).......)).)))))......((((((.......))).))))---))).............-- ( -28.91) >DroSim_CAF1 101439 105 - 1 AAGAGCGUGCCACUGGAACAAUGCCAAGUCCAGAUGGACUGGAUU-GUUGGAGAAUUGAGUGGAAAACACUAAGGAAAACUACCUACAGA---UGCGAAGUACUCACAU-- ..(((..(((..(((.((((((.(((.((((....))))))))))-)))((.......((((.....)))).((.....)).))..))).---.))).....)))....-- ( -26.20) >DroEre_CAF1 103279 93 - 1 UAGAGCGUGCCACUGGAACAAUGCCAAGUCCAGAUCAACUGGAUUUAGUGGAUAGUUGAGUGGAAAACACUGAGGGAAACCACUUAAAA----AGUG-------------- ...((.((.(((((.((....(((.((((((((.....)))))))).))).....)).)))))...)).))..((....))........----....-------------- ( -27.80) >DroYak_CAF1 103106 94 - 1 AAGAGCGUGCCACUGGCACAAUGCCAAGUCCAGAUCGACUGGAUUUAGU------------GGAAAACACUGAGGAAAACUACCUAAAA----GGUGAAGC-UUGACAUUA ..((((...((((((((.....)))((((((((.....)))))))))))------------))....((((.(((.......)))....----))))..))-))....... ( -30.90) >DroAna_CAF1 97676 97 - 1 GAGAGCGUGCCACUGGAACAAUGCCAAGUCCAGAAGGACGGGAUUC---GGUCAACAGAGUGGGGUGCACUUGGGGAAAUCCCAUAGACACCCUGCGAG---------U-- ..((.((..((..(((.......))).((((....)))).))...)---).))......(..(((((....((((.....))))....)))))..)...---------.-- ( -33.70) >consensus AAGAGCGUGCCACUGGAACAAUGCCAAGUCCAGAUGGACUGGAUU_GGUGGAGAAUUGAGUGGAAAACACUGAGGAAAACCACCUAAAGA___UGCGAAGU_CUGACAU__ ....((((.(((((((.......)).(((((((.....))))))).............)))))...)).....((....)).............))............... (-15.53 = -15.45 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:32 2006