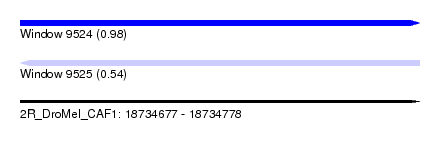
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,734,677 – 18,734,778 |
| Length | 101 |
| Max. P | 0.975088 |

| Location | 18,734,677 – 18,734,778 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 74.61 |
| Mean single sequence MFE | -17.02 |
| Consensus MFE | -3.58 |
| Energy contribution | -3.30 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.21 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18734677 101 + 20766785 -CCUUU-CCCACAUAUGCAAAUA--UG--------U--AUACGUGGAGUAGAGA-UUUUUUCCACACAAAGAA--AAAUAUAAUUUUGGAACAAUCAUUUUCUCUGCCCCUUCACUAA -.....-.((((.((((((....--))--------)--))).)))).(((((((-.((((((........)))--)))..........((....))....)))))))........... ( -20.30) >DroSec_CAF1 97457 111 + 1 -CCUUU-CCCACAUAUUCAUACAUACGUAUAUGCAUACAUACGUUGGGUA-UGA-UUUUUUCCACACAAAUAA--CAA-AUAAUUUGUGAACAAUCAUUUUCUCUGCCCUUUCGCUAA -.....-.........((((((..((((((........))))))...)))-)))-.........(((((((..--...-...)))))))................((......))... ( -18.00) >DroSim_CAF1 98263 101 + 1 -CCUUU-UCAACAUAUGCAAAUA--UG--------UAUAUACGUGGAGAA-UGA-UUUUUUCCACACAAAUAA--CAA-AUAAUUUGUGAACAAUCAUUUUCUCUGCCCUUUCGCUAA -.....-.....(((((((....--))--------)))))..(..(((((-(((-((.((....(((((((..--...-...))))))))).))))))..))))..)........... ( -19.80) >DroEre_CAF1 98659 106 + 1 UUAUUUGCAAAUAUAUGCAUAUC--UA--------CAUAUAUGUAGAAUA-CGAUACGAUCCCACACAAAUAGCACGA-AUAAUUUGCGAACAAUGAUUUUCUCCGCCCUUUUACUAA ...(((((((((...(((...((--((--------((....))))))...-.(((...)))...........)))...-...)))))))))........................... ( -13.60) >DroYak_CAF1 99707 98 + 1 GUAUUU-CACACAUAUGCAAAUU--U------------GUAUGUAGAAUA-UGA-AUUUUUCCACACAAGUAA--CAA-AUAAUUUGUGAACAAUGAUUUUCUCUGCCCUUUUACUAA (((...-.......(((((....--)------------))))(((((...-.((-(((......(((((((..--...-...)))))))......)))))..))))).....)))... ( -13.40) >consensus _CCUUU_CCCACAUAUGCAAAUA__UG________UA_AUACGUAGAGUA_UGA_UUUUUUCCACACAAAUAA__CAA_AUAAUUUGUGAACAAUCAUUUUCUCUGCCCUUUCACUAA ..........................................((((((....(........)..(((((((...........)))))))....................))))))... ( -3.58 = -3.30 + -0.28)

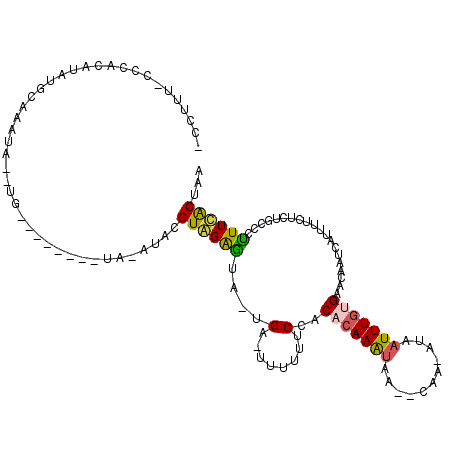
| Location | 18,734,677 – 18,734,778 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.61 |
| Mean single sequence MFE | -22.72 |
| Consensus MFE | -7.62 |
| Energy contribution | -7.46 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18734677 101 - 20766785 UUAGUGAAGGGGCAGAGAAAAUGAUUGUUCCAAAAUUAUAUUU--UUCUUUGUGUGGAAAAAA-UCUCUACUCCACGUAU--A--------CA--UAUUUGCAUAUGUGGG-AAAGG- ...........(((((((((((((((.......))))))...)--))))))))(((((.....-..)))))(((((((((--.--------((--....)).)))))))))-.....- ( -24.80) >DroSec_CAF1 97457 111 - 1 UUAGCGAAAGGGCAGAGAAAAUGAUUGUUCACAAAUUAU-UUG--UUAUUUGUGUGGAAAAAA-UCA-UACCCAACGUAUGUAUGCAUAUACGUAUGUAUGAAUAUGUGGG-AAAGG- ...((......)).......((((((.((((((...((.-...--.))....))))))...))-)))-).((((.(((((.((((((((....)))))))).)))))))))-.....- ( -28.70) >DroSim_CAF1 98263 101 - 1 UUAGCGAAAGGGCAGAGAAAAUGAUUGUUCACAAAUUAU-UUG--UUAUUUGUGUGGAAAAAA-UCA-UUCUCCACGUAUAUA--------CA--UAUUUGCAUAUGUUGA-AAAGG- ((((((.....(((((((.(((((((.((((((...((.-...--.))....))))))...))-)))-)).))...((....)--------).--..)))))...))))))-.....- ( -18.70) >DroEre_CAF1 98659 106 - 1 UUAGUAAAAGGGCGGAGAAAAUCAUUGUUCGCAAAUUAU-UCGUGCUAUUUGUGUGGGAUCGUAUCG-UAUUCUACAUAUAUG--------UA--GAUAUGCAUAUAUUUGCAAAUAA .........((((((.(.....).))))))(((((((((-.((((((((.((((((((((((...))-.))))))))))...)--------))--).)))).))).))))))...... ( -26.10) >DroYak_CAF1 99707 98 - 1 UUAGUAAAAGGGCAGAGAAAAUCAUUGUUCACAAAUUAU-UUG--UUACUUGUGUGGAAAAAU-UCA-UAUUCUACAUAC------------A--AAUUUGCAUAUGUGUG-AAAUAC ..((((...((((((.(.....).))))))((((.....-)))--)))))(((((((((....-...-..))))))))).------------.--..((..((....))..-)).... ( -15.30) >consensus UUAGUGAAAGGGCAGAGAAAAUGAUUGUUCACAAAUUAU_UUG__UUAUUUGUGUGGAAAAAA_UCA_UACUCCACGUAU_UA________CA__UAUUUGCAUAUGUGGG_AAAGG_ ...(((((.((((((.........))))))....................((((((((.............))))))))..................)))))................ ( -7.62 = -7.46 + -0.16)

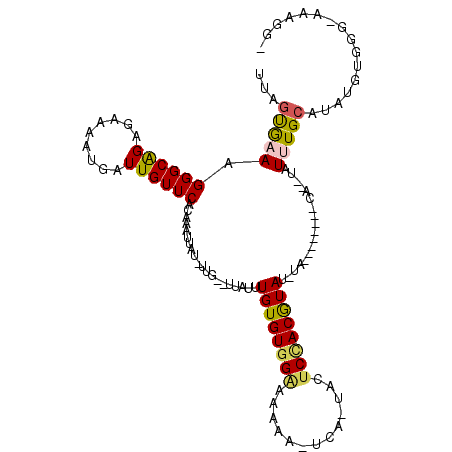
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:31 2006