| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,720,207 – 18,720,309 |
| Length | 102 |
| Max. P | 0.513168 |

| Location | 18,720,207 – 18,720,309 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 74.58 |
| Mean single sequence MFE | -24.03 |
| Consensus MFE | -7.65 |
| Energy contribution | -8.91 |
| Covariance contribution | 1.26 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.32 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
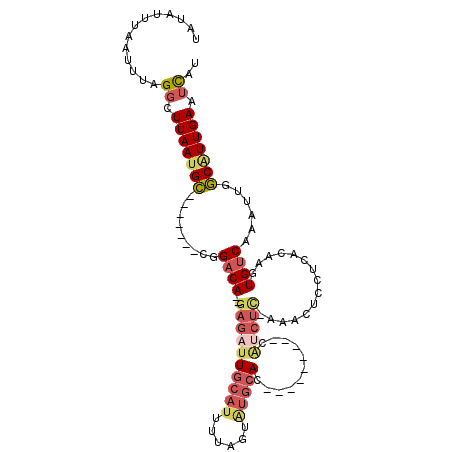
>2R_DroMel_CAF1 18720207 102 - 20766785 UAUAUUUAAUUUAGGCUUAAUGC-------CGGACA--GAGAUUGCAUUUUAGUAUGCACCCGCGCCGCCAUCUC-AAAUUCCUCACAAGUGUCAAAUUGGCCUUGAAUCAU .............(((.....))-------)(((..--((((((((((......)))))...((...)).)))))-....)))...((((.(((.....)))))))...... ( -21.30) >DroGri_CAF1 122838 98 - 1 UGUGUUUAAUUCAGCAUUAAUGAAACAUACACGUCACAGAGAUUUU---UUGUUCUUCAU---------UUACUU-GUUGUACACCAAAGUGCC-AAUUGUCGUUGAAUUAU ......(((((((((...(((((((((.....(((.....)))...---.)))..)))))---------).....-...((((......)))).-.......))))))))). ( -15.90) >DroSec_CAF1 83502 92 - 1 UAUAUUUAAUUUAGGCUUAAUGC-------CGGACA--GAGAUUGCAUUUUAGUAUGCAC---------CAUCUC-AAACUCCUC-CAAGUGUCAAAUUGGCAUUGAGUCAU .............((((((((((-------(((((.--((((((((((......))))).---------.)))))-..(((....-..))))))....)))))))))))).. ( -29.90) >DroSim_CAF1 84033 93 - 1 UAUAUUUAAUUUAGGCUUAAUGC-------CGGACA--GAGAUUGCAUUUUAGUAUGCAC---------CAUCUC-AAACUCCCCGCAAGUGUCAAAUUGGCAUUGAGUCAU .............((((((((((-------((((((--((((((((((......))))).---------.)))))-........(....)))))....)))))))))))).. ( -31.70) >DroEre_CAF1 84488 102 - 1 UAUAUUUAAUUUAGGCUUAAUGC-------CGGACA--GAGAUUGCAUUUUAGUAUGCACCC-CGCCGUCGUCUCCCACCUACACACAAGUGUCAAAUUGGCAUUGAAUCAU .............(..(((((((-------(((...--((((((((((......)))))...-.......)))))...)).((((....))))......))))))))..).. ( -24.40) >DroYak_CAF1 84900 99 - 1 UAUAUUUAAUUUAGGCUUAAUGC-------CGGACA--GAGAUUGCAUUUUAGUGUGCACCCCCGCCGCCG----CCGCCUCCUCAAAAGUGUCAAAUUGGCAUUGAAUCAU .............(..(((((((-------((((((--(((..(((((......))))).....((....)----)......))).....))))....)))))))))..).. ( -21.00) >consensus UAUAUUUAAUUUAGGCUUAAUGC_______CGGACA__GAGAUUGCAUUUUAGUAUGCAC_________CAUCUC_AAACUCCUCACAAGUGUCAAAUUGGCAUUGAAUCAU .............((.(((((((.........((((..((((((((((......)))))...........)))))...............))))......))))))).)).. ( -7.65 = -8.91 + 1.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:26 2006