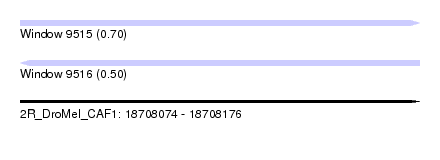
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,708,074 – 18,708,176 |
| Length | 102 |
| Max. P | 0.699157 |

| Location | 18,708,074 – 18,708,176 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.33 |
| Mean single sequence MFE | -35.37 |
| Consensus MFE | -24.29 |
| Energy contribution | -26.23 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
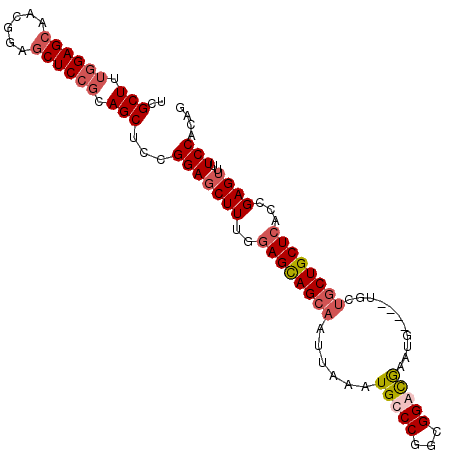
>2R_DroMel_CAF1 18708074 102 + 20766785 UCGCUUUGGAGCUCCGCAACUCCGCAGCUCCGGACCUUUGGAGUAGCAAUUAAAUGCCCGGCGGAUGAAUAUGUGCGCUGCUGCUCACCGAGUUUUCCACAG .......((((((.((......)).))))))(((.(((.(((((((((............(((.((......)).))))))))))).).)))...))).... ( -31.20) >DroSec_CAF1 71547 98 + 1 UCGCUUUGGAGCAACGGAGCUCCGCAGCUUCGGAGCUUUGAAGCAGCAAUUAAAUGCCCGGCGGGCGAAUG----UGCUGCUGCUCACCGAGUUUUCCACAG ..((((.(((((..((((((((((......)))))))))).(((((((......(((((...)))))....----))))))))))).).))))......... ( -42.40) >DroSim_CAF1 71963 98 + 1 UCGCUUUGGAGCAACGGAGCUCCGCAGCUCCGGAGCUUUGGAGCAGCAAUUAAAUGCCCGGCGGGCGAAUG----UGCUGCUGCUCACCGAGUUUUCCACAG ..((((((((((..(((....)))..))))))))))..((((((((((......(((((...)))))....----)))))).((((...))))..))))... ( -43.90) >DroEre_CAF1 72478 79 + 1 UCGCUUUG----------------CAGCUUCGGAGCUUUGGAGCAGCAAUUAAAUGCCCGGCGGACAAAUG----UG---CUGCUCACCGAGUUUUCCACAG ..(((...----------------.)))...(((((((.(((((((((........((....)).......----))---)))))).).))))..))).... ( -23.96) >consensus UCGCUUUGGAGCAACGGAGCUCCGCAGCUCCGGAGCUUUGGAGCAGCAAUUAAAUGCCCGGCGGACGAAUG____UGCUGCUGCUCACCGAGUUUUCCACAG ..(((.((((((......)))))).)))...(((((((..((((((((......(((((...)))))...........))))))))...))))..))).... (-24.29 = -26.23 + 1.94)

| Location | 18,708,074 – 18,708,176 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.33 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -24.42 |
| Energy contribution | -25.61 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
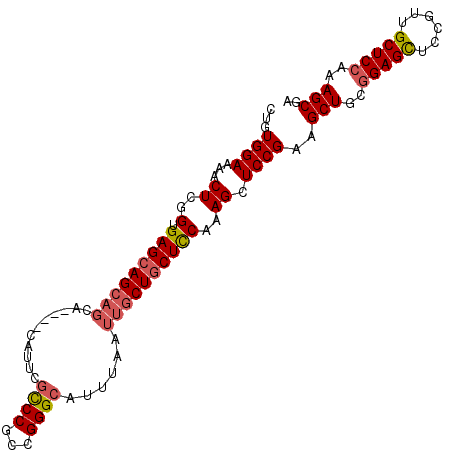
>2R_DroMel_CAF1 18708074 102 - 20766785 CUGUGGAAAACUCGGUGAGCAGCAGCGCACAUAUUCAUCCGCCGGGCAUUUAAUUGCUACUCCAAAGGUCCGGAGCUGCGGAGUUGCGGAGCUCCAAAGCGA ..(((((.......(((.((....)).))).......)))))((((((......)))).............((((((.((......)).))))))....)). ( -30.94) >DroSec_CAF1 71547 98 - 1 CUGUGGAAAACUCGGUGAGCAGCAGCA----CAUUCGCCCGCCGGGCAUUUAAUUGCUGCUUCAAAGCUCCGAAGCUGCGGAGCUCCGUUGCUCCAAAGCGA ((.((((.....(((((((((((((..----.....((((...))))......))))))).))).(((((((......))))))))))....)))).))... ( -36.32) >DroSim_CAF1 71963 98 - 1 CUGUGGAAAACUCGGUGAGCAGCAGCA----CAUUCGCCCGCCGGGCAUUUAAUUGCUGCUCCAAAGCUCCGGAGCUGCGGAGCUCCGUUGCUCCAAAGCGA (((.(.....).))).(((((((((..----.....((((...))))......)))))))))....(((..(((((.((((....)))).)))))..))).. ( -40.82) >DroEre_CAF1 72478 79 - 1 CUGUGGAAAACUCGGUGAGCAG---CA----CAUUUGUCCGCCGGGCAUUUAAUUGCUGCUCCAAAGCUCCGAAGCUG----------------CAAAGCGA ...((((...((..(.((((((---((----....(((((...)))))......)))))))))..)).))))..(((.----------------...))).. ( -25.50) >consensus CUGUGGAAAACUCGGUGAGCAGCAGCA____CAUUCGCCCGCCGGGCAUUUAAUUGCUGCUCCAAAGCUCCGAAGCUGCGGAGCUCCGUUGCUCCAAAGCGA ...((((...((..(.(((((((((...........((((...))))......))))))))))..)).))))..(((..(((((......)))))..))).. (-24.42 = -25.61 + 1.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:22 2006