
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,696,652 – 18,696,757 |
| Length | 105 |
| Max. P | 0.525733 |

| Location | 18,696,652 – 18,696,757 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.77 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -14.98 |
| Energy contribution | -15.46 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18696652 105 + 20766785 ---AACAUUAAAAUGCACUUGUUAACAAGUAAUUUGUUGG--GAAUGGCAGAUAUUUGCUUAUCGCAGCCAUGAAAUCA---------GUUUAGCCGACUA-AAAAAAGGUUGAAUAUUU ---..((...((((..(((((....))))).))))..)).--..(((((.((((......))))...))))).......---------((((((((.....-......)))))))).... ( -22.80) >DroVir_CAF1 88719 108 + 1 GCUCGCAUUAAAAUGCACUUGUUAACAAGUAAUUUGUCCG-GCAAUGGCAGAUAUUUGCUUAUCGCAGUCAUGAAAUCA---------GUUCAGCGAAAUA-AA-AAAGGUUGAAUAUUU (((.((((....))))(((((....))))).........)-)).(((((.((((......))))...))))).......---------(((((((......-..-....))))))).... ( -21.80) >DroPse_CAF1 66578 104 + 1 ---AACAUUAAAAUGCACUUGUUAACAAGUAAUUUGUUGGUGGAAUGGCAGAUAUUUGCUUAUCGCAGCCAUGAAAUCACGGUCUGCCGUUUAUCC-------------GUUGAAUAUUU ---.............(((((....))))).((((..(((..((((((((((...((((.....))))((.((....)).))))))))))))..))-------------)..)))).... ( -26.50) >DroWil_CAF1 81828 109 + 1 UUUCGCAUUAAAAUGCACUUGUUAACAAGUAAUUUGUUGG-GGAAUGGCUGAUAUUUGCUUAUCGCCGUCAUGAAAUCA---------GUUCAAACGAAACCCA-GUAGGUUGAAUAUUU .(((((((....))))(((((....)))))((((((((((-(((.((((.((((......)))))))))).((((....---------.)))).......))))-))))))))))..... ( -27.30) >DroAna_CAF1 60426 101 + 1 ---AACAUUAAAAUGCACUUGUUAACAAGUAAUUUGUUGG--AAAUGGCAGAUAUUUGCUUAUCGCAGCCAGG-GAUCA---------GUUUAGCCGGCCG-A---GAGGUUGAAUAUUU ---..((...((((..(((((....))))).))))..))(--(..((((.((((......))))...))))..-..)).---------((((((((.....-.---..)))))))).... ( -23.00) >DroPer_CAF1 67358 104 + 1 ---AACAUUAAAAUGCACUUGUUAACAAGUAAUUUGUUGGUGGAAUGGCAGAUAUUUGCUUAUCGCAGCCAUGAAAUCACGGUCUGCCGUUUAUCC-------------GUUGAAUAUUU ---.............(((((....))))).((((..(((..((((((((((...((((.....))))((.((....)).))))))))))))..))-------------)..)))).... ( -26.50) >consensus ___AACAUUAAAAUGCACUUGUUAACAAGUAAUUUGUUGG_GGAAUGGCAGAUAUUUGCUUAUCGCAGCCAUGAAAUCA_________GUUUAGCCGA____A____AGGUUGAAUAUUU .....((...((((..(((((....))))).))))..)).....(((((.((((......))))...)))))................(((((((..............))))))).... (-14.98 = -15.46 + 0.47)

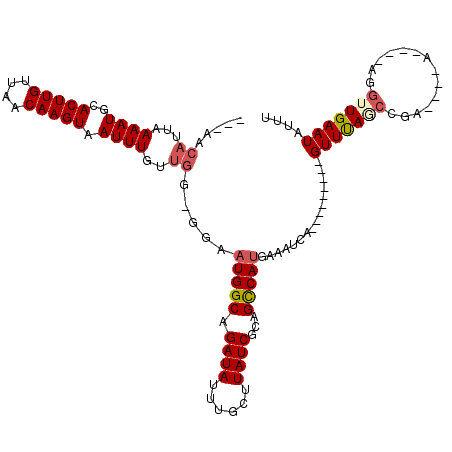
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:19 2006