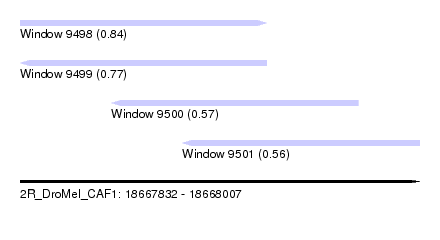
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,667,832 – 18,668,007 |
| Length | 175 |
| Max. P | 0.844225 |

| Location | 18,667,832 – 18,667,940 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.28 |
| Mean single sequence MFE | -36.37 |
| Consensus MFE | -23.21 |
| Energy contribution | -23.52 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18667832 108 + 20766785 AUGGCGUCAAUGCCUACCAGAAGGCAAUAGACAUUUACUUGAGCCAGGGACAUCAGAUACCGAUUGAGUGGCUCAAUAACUUGGCCAGCAGUCAGCUGAUGGCCAAAA .(((((((..(((((......)))))...))).....((((...))))..((((((.....(((((..((((.(((....))))))).)))))..))))))))))... ( -36.50) >DroSec_CAF1 31458 108 + 1 AAGUCGUCAAUGCCUACCAGAAGGCAAUAGACAUUUACUUGAGCCGGGGACAUCAGAUACCGAUUGAGUGGCUCAAUAACCUGGCCAACAGUCAGCUGAUGGCCAAAA .....(((..(((((......)))))...)))......((((((((....).((((.......))))..))))))).....((((((.(((....))).))))))... ( -36.80) >DroSim_CAF1 31742 108 + 1 AAGUCGUCAAUGCCUACCAGAAGGCAAUGGACAUUUACUUGAGCCGGGGACAUCAGAUACCGAUUGAGUGGCUCAAUAACCUGGCCAACAGUCAGCUGAUGGCCAAAA .....(((..(((((......)))))...)))......((((((((....).((((.......))))..))))))).....((((((.(((....))).))))))... ( -37.90) >DroEre_CAF1 31020 108 + 1 AGGCCGUCAAUGCCUACCAGAAGGCAAUUUCCAUUUACUCUGGCGAGGGACAUCACAUACCGAUUGAGUGGCUCAAUAACCUGGCCAACAGCCAACUGAUGGCCAAAA .((((((((.(((((......)))))..............((((.((((....)........((((((...))))))..))).)))).........)))))))).... ( -36.10) >DroYak_CAF1 31811 108 + 1 AGACCGUCAAUGCCUACCAGAAGGCAAUAUCCAUUUGCUCGGUUCAGGGACAUCACAUACCGAUUGAGUGGCUCAACAACCUGGCCAACAGCCAACUGAUGGCCAAAA ..........(((((......)))))....((((((..(((((.....(....)....)))))..))))))..........((((((.(((....))).))))))... ( -32.10) >DroAna_CAF1 30910 108 + 1 AGGCCAUCGAUUCCUACCAAAAGGCCAUGGCCAUUUGCGAGAAAUCGGGUCGAGAGGUGCCCGUCGAGUGGGUCAACAAUCUGGCAGCCACCCAGCAGCUGGCCAAGA .((((.................)))).((((((.((((((....))(((((.(((..((((((.....)))).))....))).).....)))).)))).))))))... ( -38.83) >consensus AGGCCGUCAAUGCCUACCAGAAGGCAAUAGACAUUUACUUGAGCCAGGGACAUCAGAUACCGAUUGAGUGGCUCAAUAACCUGGCCAACAGCCAGCUGAUGGCCAAAA .((((((((((((((......)))).....................((...........)).))))).)))))........((((((.(((....))).))))))... (-23.21 = -23.52 + 0.31)


| Location | 18,667,832 – 18,667,940 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.28 |
| Mean single sequence MFE | -39.37 |
| Consensus MFE | -23.33 |
| Energy contribution | -25.28 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18667832 108 - 20766785 UUUUGGCCAUCAGCUGACUGCUGGCCAAGUUAUUGAGCCACUCAAUCGGUAUCUGAUGUCCCUGGCUCAAGUAAAUGUCUAUUGCCUUCUGGUAGGCAUUGACGCCAU .((((((((.(((....))).))))))))...((((((((....(((((...))))).....))))))))......(((...(((((......)))))..)))..... ( -40.60) >DroSec_CAF1 31458 108 - 1 UUUUGGCCAUCAGCUGACUGUUGGCCAGGUUAUUGAGCCACUCAAUCGGUAUCUGAUGUCCCCGGCUCAAGUAAAUGUCUAUUGCCUUCUGGUAGGCAUUGACGACUU ...((((((.(((....))).))))))((((.(((((((.....(((((...)))))......)))))))......(((...(((((......)))))..))))))). ( -39.20) >DroSim_CAF1 31742 108 - 1 UUUUGGCCAUCAGCUGACUGUUGGCCAGGUUAUUGAGCCACUCAAUCGGUAUCUGAUGUCCCCGGCUCAAGUAAAUGUCCAUUGCCUUCUGGUAGGCAUUGACGACUU ...((((((.(((....))).))))))((((.(((((((.....(((((...)))))......)))))))......(((...(((((......)))))..))))))). ( -38.50) >DroEre_CAF1 31020 108 - 1 UUUUGGCCAUCAGUUGGCUGUUGGCCAGGUUAUUGAGCCACUCAAUCGGUAUGUGAUGUCCCUCGCCAGAGUAAAUGGAAAUUGCCUUCUGGUAGGCAUUGACGGCCU .((((((((.(((....))).))))))))..((((((...)))))).(((...(((((((....(((((((..(((....)))...))))))).)))))))...))). ( -38.10) >DroYak_CAF1 31811 108 - 1 UUUUGGCCAUCAGUUGGCUGUUGGCCAGGUUGUUGAGCCACUCAAUCGGUAUGUGAUGUCCCUGAACCGAGCAAAUGGAUAUUGCCUUCUGGUAGGCAUUGACGGUCU ...((((((.(((....))).))))))(..(((..((((.....(((((...(..((((((.((.......))...))))))..)...))))).))).)..)))..). ( -37.10) >DroAna_CAF1 30910 108 - 1 UCUUGGCCAGCUGCUGGGUGGCUGCCAGAUUGUUGACCCACUCGACGGGCACCUCUCGACCCGAUUUCUCGCAAAUGGCCAUGGCCUUUUGGUAGGAAUCGAUGGCCU ....(((((.....((((.((.((((....((((((.....)))))))))))).))))...(((((((((.((((.(((....))).))))).)))))))).))))). ( -42.70) >consensus UUUUGGCCAUCAGCUGACUGUUGGCCAGGUUAUUGAGCCACUCAAUCGGUAUCUGAUGUCCCUGGCUCAAGUAAAUGGCUAUUGCCUUCUGGUAGGCAUUGACGGCCU ..(((((((.(((....))).)))))))(((((((((((.....((((.....))))......)))))))............(((((......))))).))))..... (-23.33 = -25.28 + 1.95)


| Location | 18,667,872 – 18,667,980 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.65 |
| Mean single sequence MFE | -40.58 |
| Consensus MFE | -30.66 |
| Energy contribution | -31.83 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18667872 108 - 20766785 GAAAGGGCAUCGUCAAGGGUAUUGAGUGCCUUCUCUGGCAUUUUGGCCAUCAGCUGACUGCUGGCCAAGUUAUUGAGCCACUCAAUCGGUAUCUGAUGUCCCUGGCUC (..((((((((.....((((((((((((((......)))))((((((((.(((....))).))))))))...(((((...)))))))))))))))))).))))..).. ( -42.00) >DroSec_CAF1 31498 108 - 1 GAAAGCGCUUCGUCAAGGGUAUCGAGUGCCCUCUCUGGCAUUUUGGCCAUCAGCUGACUGUUGGCCAGGUUAUUGAGCCACUCAAUCGGUAUCUGAUGUCCCCGGCUC ......(((.(((((..(((((((((((((......)))))..((((((.(((....))).))))))((((....))))......))))))))))))).....))).. ( -36.70) >DroSim_CAF1 31782 108 - 1 GAAAGCGCUUCGUCAAGGGUAUCGAGUGCCCUCUCUGGCAUUUUGGCCAUCAGCUGACUGUUGGCCAGGUUAUUGAGCCACUCAAUCGGUAUCUGAUGUCCCCGGCUC ......(((.(((((..(((((((((((((......)))))..((((((.(((....))).))))))((((....))))......))))))))))))).....))).. ( -36.70) >DroEre_CAF1 31060 108 - 1 GAAAGGGCAUCGUCAAGGGUAUCGAGUGCCUGCUCUGGCAUUUUGGCCAUCAGUUGGCUGUUGGCCAGGUUAUUGAGCCACUCAAUCGGUAUGUGAUGUCCCUCGCCA (..((((...(((((...((((((((((((......)))))..((((((.(((....))).))))))((((....))))......))))))).))))).))))..).. ( -42.00) >DroYak_CAF1 31851 108 - 1 GAAAGGGCAUCGUCGAGGGUUUCGAGUGCCUGCGCUGGCAUUUUGGCCAUCAGUUGGCUGUUGGCCAGGUUGUUGAGCCACUCAAUCGGUAUGUGAUGUCCCUGAACC ...(((((((((((((((((...(((((((......)))))))((((((.(((....))).))))))((((....))))))))..))))....))))).))))..... ( -41.40) >DroAna_CAF1 30950 108 - 1 GCCAGGGCCUCGUCGAUGGUGGCGAGGGCCUGCUGCGGCAUCUUGGCCAGCUGCUGGGUGGCUGCCAGAUUGUUGACCCACUCGACGGGCACCUCUCGACCCGAUUUC ((.(((.((((((((....)))))))).)))))..(((((..(((((.(((..(...)..))))))))..)))))..........(((((.......).))))..... ( -44.70) >consensus GAAAGGGCAUCGUCAAGGGUAUCGAGUGCCUGCUCUGGCAUUUUGGCCAUCAGCUGACUGUUGGCCAGGUUAUUGAGCCACUCAAUCGGUAUCUGAUGUCCCUGGCUC ....(((...(((((..(((((((((((((......))))))(((((((.(((....))).)))))))...((((((...)))))))))))))))))).)))...... (-30.66 = -31.83 + 1.17)

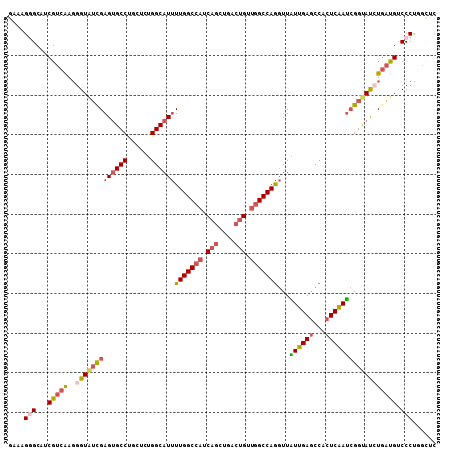
| Location | 18,667,903 – 18,668,007 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 80.26 |
| Mean single sequence MFE | -36.52 |
| Consensus MFE | -22.78 |
| Energy contribution | -22.95 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18667903 104 - 20766785 UUAUCGCUAUUCAUUACU-CUACAUUUGGAAAGGGCAUCGUCAAGGGUAUUGAGUGCCUUCUCUGGCAUUUUGGCCAUCAGCUGACUGCUGGCCAAGUUAUUGAG ................((-(.....(..((.(((((((..((((.....))))))))))).))..)...((((((((.(((....))).)))))))).....))) ( -30.60) >DroSec_CAF1 31529 104 - 1 UGUUCGCUAUUCAAUACU-CUACAUUUGGAAAGCGCUUCGUCAAGGGUAUCGAGUGCCCUCUCUGGCAUUUUGGCCAUCAGCUGACUGUUGGCCAGGUUAUUGAG ....((((.(((((....-......))))).))))....(((((((((((...)))))))...))))..((((((((.(((....))).))))))))........ ( -32.70) >DroSim_CAF1 31813 104 - 1 UGUUCGCUAUUCAAUACU-CUACAUUUGGAAAGCGCUUCGUCAAGGGUAUCGAGUGCCCUCUCUGGCAUUUUGGCCAUCAGCUGACUGUUGGCCAGGUUAUUGAG ....((((.(((((....-......))))).))))....(((((((((((...)))))))...))))..((((((((.(((....))).))))))))........ ( -32.70) >DroEre_CAF1 31091 104 - 1 UGCUCCCCAUUCAGUGUU-CUACAUUUGGAAAGGGCAUCGUCAAGGGUAUCGAGUGCCUGCUCUGGCAUUUUGGCCAUCAGUUGGCUGUUGGCCAGGUUAUUGAG (((((.(((...((((..-...)))))))...)))))...((((....((((((((((......)))))))((((((.(((....))).)))))))))..)))). ( -34.70) >DroYak_CAF1 31882 104 - 1 UGAUCGCCAUUCGAUGUU-CUGCACUUGGAAAGGGCAUCGUCGAGGGUUUCGAGUGCCUGCGCUGGCAUUUUGGCCAUCAGUUGGCUGUUGGCCAGGUUGUUGAG .(((((((.(((((((..-.(((.(((....)))))).))))))))))...(((((((......)))))))((((((.(((....))).))))))))))...... ( -41.50) >DroAna_CAF1 30981 104 - 1 UGCUCGUCC-CCAGCCCUGUCGCUCCUGGCCAGGGCCUCGUCGAUGGUGGCGAGGGCCUGCUGCGGCAUCUUGGCCAGCUGCUGGGUGGCUGCCAGAUUGUUGAC .((..((((-(((((..........((((((((((((((((((....))))))))(((......))).))))))))))..)))))).))).))............ ( -46.90) >consensus UGAUCGCCAUUCAAUACU_CUACAUUUGGAAAGGGCAUCGUCAAGGGUAUCGAGUGCCUGCUCUGGCAUUUUGGCCAUCAGCUGACUGUUGGCCAGGUUAUUGAG ..........((((((.......................((((.(((((.....)))))....))))...(((((((.(((....))).)))))))..)))))). (-22.78 = -22.95 + 0.17)

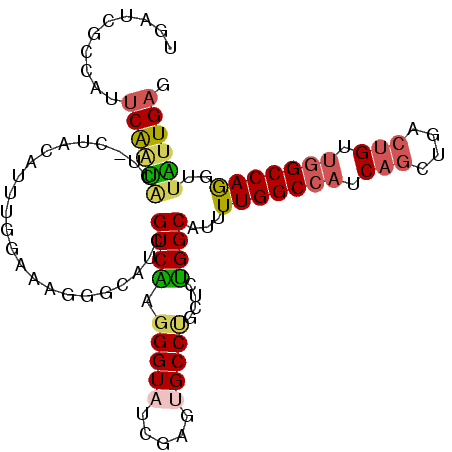
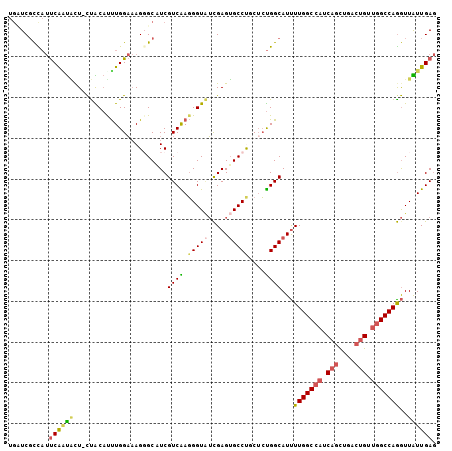
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:07 2006