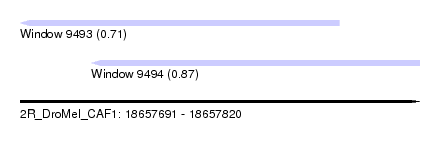
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,657,691 – 18,657,820 |
| Length | 129 |
| Max. P | 0.868402 |

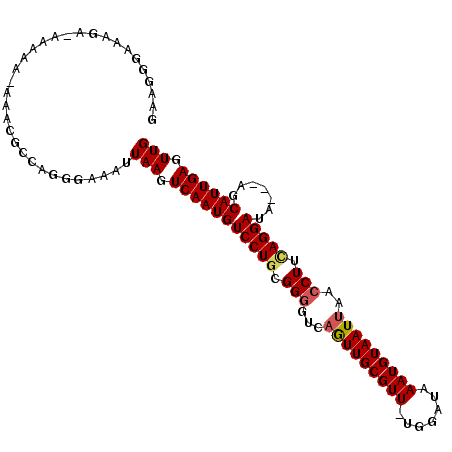
| Location | 18,657,691 – 18,657,794 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 84.88 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -22.98 |
| Energy contribution | -24.87 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18657691 103 - 20766785 GAAAUUAAGUCAAUGUCCUGCGGGGUCAGUUGCGUUUUGGAUAAAUGUAAUUAACCUUUAGGAUCGCGAGCAUUGAGUUGGGUAUCCCAACUUAAACAACGGA----- .........((...((((((.(((((.((((((((((.....)))))))))).)))))))))))...))...((((((((((...))))))))))........----- ( -31.70) >DroSec_CAF1 21228 103 - 1 GAAAUUAAGUCAAUGUCCUGCGGGGUCAGUUGCGUUUUGGAUAAAUGUAAUUAACCUUCAGGAUCGCGAGCAUUGAGUUGGGUAGCCCAACUUAAACAACGCA----- ..............((((((.(((((.((((((((((.....)))))))))).))))))))))).(((....((((((((((...))))))))))....))).----- ( -37.60) >DroEre_CAF1 21208 103 - 1 GAAAUUAAGUCAAUGUCCUGCGGGUUCAGUUGCGUUUUGGAUAAAUGUAAUUAACCUUCAGGAUCGCCAGCAUUGAGUUGGGUAGCCCAACUUAAACAGCGGA----- ..............((((((.(((((.((((((((((.....))))))))))))))).))))))(((.....((((((((((...))))))))))...)))..----- ( -36.80) >DroWil_CAF1 32262 100 - 1 GAAAUUAAGUCAAUGUCCUGCGGGGUCAGUUGCGUU-UGGAUAAAUGUAAUUAACCUACAGGAUA---AGCAUUGAGUUGGGUCCU----UUUAAUAAUCAAGGACAA ....((((.(((((((((((..((((.(((((((((-(....)))))))))).)))).))))...---.))))))).))))(((((----(.........)))))).. ( -33.30) >DroYak_CAF1 21653 103 - 1 GAAAUUAAGCCAAUGUCCUGCGGGGUCAGUUGCGUUUUGGAUAAAUGUAAUUAACCUUCAGGAUCGCGCGAGUUGAGUUGGGUAGUCCAACUUAAACAACGGA----- ........((....((((((.(((((.((((((((((.....)))))))))).))))))))))).)).((..(((((((((.....)))))))))....))..----- ( -33.60) >DroAna_CAF1 21844 95 - 1 GAAAUUAAGUCAAUGUCCUGCGG-GUCAGUUGCGUUUUGGAUAAAUGUAAUUAACCUUAAGGAUCGACGGCAUUGAGUUGGGUCGCC-------AGGGACGGA----- ....((((.((((((((((..((-((.((((((((((.....)))))))))).))))..))).......))))))).))))(((...-------...)))...----- ( -26.71) >consensus GAAAUUAAGUCAAUGUCCUGCGGGGUCAGUUGCGUUUUGGAUAAAUGUAAUUAACCUUCAGGAUCGCGAGCAUUGAGUUGGGUAGCCCAACUUAAACAACGGA_____ ..............((((((.(((...((((((((((.....))))))))))..))).))))))........((((((((((...))))))))))............. (-22.98 = -24.87 + 1.89)

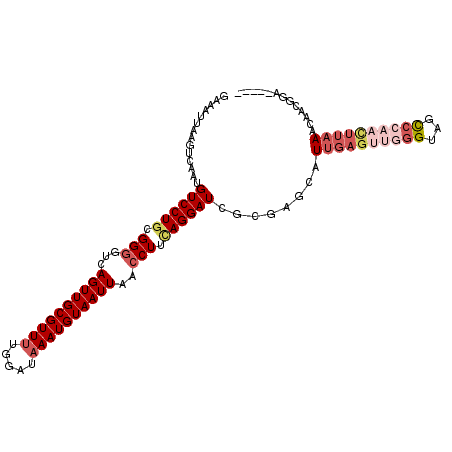
| Location | 18,657,714 – 18,657,820 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.55 |
| Mean single sequence MFE | -26.39 |
| Consensus MFE | -20.29 |
| Energy contribution | -20.54 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18657714 106 - 20766785 GACGAUGAAGA-ACAAC-AAACGCCACGGAAAUUAAGUCAAUGUCCUGCGGGGUCAGUUGCGUUUUGGAUAAAUGUAAUUAACCUUUAGGAUCGCGAGCAUUGAGUUG ...........-.((((-.....(....)........(((((((((((.(((((.((((((((((.....)))))))))).)))))))))).(....))))))))))) ( -24.90) >DroVir_CAF1 43340 103 - 1 GGAGGGGUGGAAAAAAA-AAACGCUGGGGAAAUUAAGUCAAUGUCCUGCGGGGUCAGUUGCGUU-UGGAUAAAUGUAAUUAACCUUCAGGAUA---AGCAUUGAGUUG .....((((........-...))))........(((.(((((((((((.(((((.(((((((((-(....)))))))))).)))))))))...---.))))))).))) ( -28.60) >DroGri_CAF1 42643 103 - 1 AAACAAAAUAAAAGAAA-AAACGCUGGGGAAAUUAAGUCAAUGUCCUGCGGGGUCAGUUGCGUU-UGGAUAAAUGUAAUUAACCUUCAGGAUA---AGCAUUGAGUUG .................-...............(((.(((((((((((.(((((.(((((((((-(....)))))))))).)))))))))...---.))))))).))) ( -26.20) >DroWil_CAF1 32286 101 - 1 UGCGGGAAAA---AAACAGAACGCCAGGGAAAUUAAGUCAAUGUCCUGCGGGGUCAGUUGCGUU-UGGAUAAAUGUAAUUAACCUACAGGAUA---AGCAUUGAGUUG .(((......---........))).........(((.(((((((((((..((((.(((((((((-(....)))))))))).)))).))))...---.))))))).))) ( -27.74) >DroAna_CAF1 21860 99 - 1 AA----A-AGG-AAAAC--AGCGCCAGGGAAAUUAAGUCAAUGUCCUGCGG-GUCAGUUGCGUUUUGGAUAAAUGUAAUUAACCUUAAGGAUCGACGGCAUUGAGUUG ..----.-...-....(--((.(((((.....))..(((...(((((..((-((.((((((((((.....)))))))))).))))..))))).)))))).)))..... ( -25.40) >DroPer_CAF1 24949 101 - 1 GAGGGUA-GG-----GA-AAACGGCAUGGAAAUUAAGUCAAUGUCCUGCGGGUUCCAUUGCGUUGUGGAUAAAUGUAAUUAACCUUCAGGAUCAGAAUCAUUGAGUUG ((((((.-((-----((-...(.(((.(((.(((.....))).)))))).).))))((((((((.......))))))))..))))))..................... ( -25.50) >consensus GAAGGGAAAGA_AAAAA_AAACGCCAGGGAAAUUAAGUCAAUGUCCUGCGGGGUCAGUUGCGUU_UGGAUAAAUGUAAUUAACCUUCAGGAUA___AGCAUUGAGUUG .................................(((.(((((((((((.(((...(((((((((.......)))))))))..))).))))).......)))))).))) (-20.29 = -20.54 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:58:00 2006