| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,656,258 – 18,656,357 |
| Length | 99 |
| Max. P | 0.954158 |

| Location | 18,656,258 – 18,656,357 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 90.45 |
| Mean single sequence MFE | -33.44 |
| Consensus MFE | -25.64 |
| Energy contribution | -26.24 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
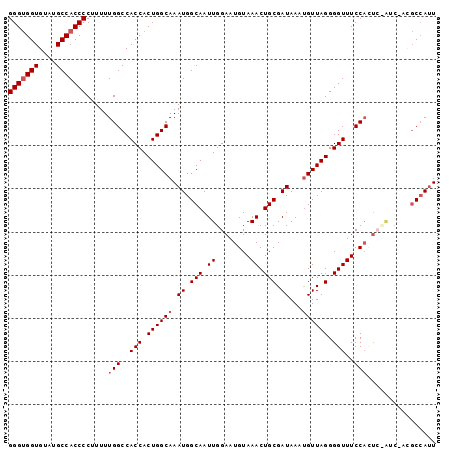
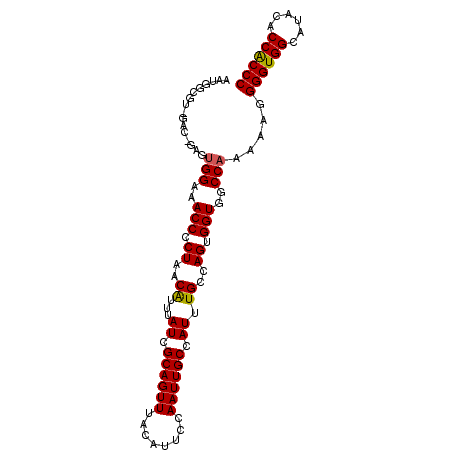
>2R_DroMel_CAF1 18656258 99 + 20766785 GGGUGGUGUAUGCCACCCCUUUUUGGCCACCACUGGCAAAUGGCAAUUGGAAUGUAAACUGCGAUAAAUGUUAGGGGUUUCCACUCCAUCCACGCCAUU (((((((....))))))).......((((....)))).((((((...((((.((.(((((.(...........).))))).)).)))).....)))))) ( -34.80) >DroSec_CAF1 19903 99 + 1 GGGUGGUGUAUGCCACCCCUUUUUGGCCACCACUGGCAAAUGGCAAUUGGAAUGUAAACUGCGAUAAAUGUUAGGGGUUUCCACUCCAUCCACGCCAUU (((((((....))))))).......((((....)))).((((((...((((.((.(((((.(...........).))))).)).)))).....)))))) ( -34.80) >DroSim_CAF1 19984 90 + 1 GGGUGGUGUAUGCCACCCCUUUUUGGCCACCACUGGCAAAUGGCAAUUGGAAUGUAAACUGCGAUAAACGUUAGGGGUUUCC---------ACGCCAUU (((((((....))))))).......((((....)))).((((((...(((((......(((((.....)).)))....))))---------).)))))) ( -30.90) >DroEre_CAF1 19760 97 + 1 GGGUGGUGUAUGCCACCCCAUUUUGGCCACCACUGGCAAAUGGCAAUUGGAAUGUAAACUGCGAUAAAUGUUAGGGGUUUCCACUCUGUC--UGCCAUU (((((((....))))))).......((((....)))).(((((((..((((.((.(((((.(...........).))))).)).))))..--))))))) ( -35.10) >DroYak_CAF1 20179 95 + 1 GGGCGGUGUAUGCCACCCCGUUUUGGCCACCACUGGCAAAUGGCAAUUGGAAUGUAAACUGCGAUAAAUGUUAGGGGUUUCCAUU--GCC--GCCAACU .(((((((.(((..(((((......((((....))))..((.(((.((........)).))).))........)))))...))))--)))--))).... ( -31.60) >consensus GGGUGGUGUAUGCCACCCCUUUUUGGCCACCACUGGCAAAUGGCAAUUGGAAUGUAAACUGCGAUAAAUGUUAGGGGUUUCCACUC_AUC_ACGCCAUU (((((((....))))))).....(((..(((.((((((.((.(((.((........)).))).))...)))))).)))..)))................ (-25.64 = -26.24 + 0.60)


| Location | 18,656,258 – 18,656,357 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 90.45 |
| Mean single sequence MFE | -29.47 |
| Consensus MFE | -22.90 |
| Energy contribution | -22.78 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18656258 99 - 20766785 AAUGGCGUGGAUGGAGUGGAAACCCCUAACAUUUAUCGCAGUUUACAUUCCAAUUGCCAUUUGCCAGUGGUGGCCAAAAAGGGGUGGCAUACACCACCC ..((((((((.(((((((.((((.................)))).)))))))....))))..))))((((((((((........))))...)))))).. ( -31.93) >DroSec_CAF1 19903 99 - 1 AAUGGCGUGGAUGGAGUGGAAACCCCUAACAUUUAUCGCAGUUUACAUUCCAAUUGCCAUUUGCCAGUGGUGGCCAAAAAGGGGUGGCAUACACCACCC ..((((((((.(((((((.((((.................)))).)))))))....))))..))))((((((((((........))))...)))))).. ( -31.93) >DroSim_CAF1 19984 90 - 1 AAUGGCGU---------GGAAACCCCUAACGUUUAUCGCAGUUUACAUUCCAAUUGCCAUUUGCCAGUGGUGGCCAAAAAGGGGUGGCAUACACCACCC ..(((.((---------((..((((((..........((((((........)))))).....((((....)))).....))))))..).))).)))... ( -26.20) >DroEre_CAF1 19760 97 - 1 AAUGGCA--GACAGAGUGGAAACCCCUAACAUUUAUCGCAGUUUACAUUCCAAUUGCCAUUUGCCAGUGGUGGCCAAAAUGGGGUGGCAUACACCACCC ..(((((--((..(((((...........)))))...((((((........))))))..)))))))((((((((((........))))...)))))).. ( -28.00) >DroYak_CAF1 20179 95 - 1 AGUUGGC--GGC--AAUGGAAACCCCUAACAUUUAUCGCAGUUUACAUUCCAAUUGCCAUUUGCCAGUGGUGGCCAAAACGGGGUGGCAUACACCGCCC ....(((--((.--....(..(((((...........((((((........)))))).....((((....))))......)))))..).....))))). ( -29.30) >consensus AAUGGCGU_GAC_GAGUGGAAACCCCUAACAUUUAUCGCAGUUUACAUUCCAAUUGCCAUUUGCCAGUGGUGGCCAAAAAGGGGUGGCAUACACCACCC ................(((..(((.((..((...((.((((((........)))))).)).))..)).)))..))).....((((((......)))))) (-22.90 = -22.78 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:58 2006