| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,652,113 – 18,652,219 |
| Length | 106 |
| Max. P | 0.567171 |

| Location | 18,652,113 – 18,652,219 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 67.28 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -5.34 |
| Energy contribution | -6.12 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.19 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
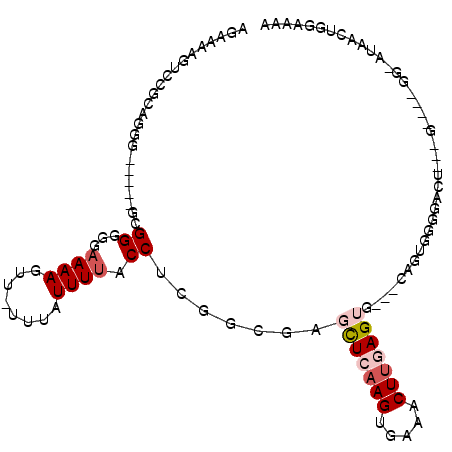
>2R_DroMel_CAF1 18652113 106 - 20766785 AGAAAAGUCCGUUGGGGAGGGGCGGGUGAAAAGUU-UUUAUUUUACCUCGGCGAGCUCAAGUGAAACUUGAGUG---CAGUUGGGACUGGGGGACUGG-GUGACUGAAAAA .....(((((.(..(........((((((((....-....))))))))((((..(((((((.....))))))).---..))))...)..).)))))((-....))...... ( -34.20) >DroVir_CAF1 36319 91 - 1 UGGCCAAUAUAAAG---------GGCA-AAAAGUUUUUUAUUUUACCUGGGCGAUUUGAAGUGAAACUAGAGCGCCAUAAUGUGAAAA---A----CC-AUAGCUGGAA-- ((.((........)---------).))-....(((((((((.(((....((((.(((..((.....)).))))))).))).)))))))---)----).-..........-- ( -17.40) >DroGri_CAF1 35285 92 - 1 UGGCCAAUAUAAAG---------GGCACAAAAGUUUUUUAUUUCACCUAGGUGAUUUGAAGUGAAACUUGAGCGCUGCAAUGUGAAAA---G----CU-AUAACUAGAA-- ((((..((((..((---------.((....((((((((((..((((....))))..)))...)))))))..)).))...)))).....---)----))-).........-- ( -18.60) >DroSec_CAF1 15850 103 - 1 AGAAAAGUCCGCAGGG---GGGCGGGGGAAAAGUU-UUUAUUUUGCCUCGGCGAGCUCAAGUGAAACUUGAGUG---CAGUUGGGACUGGGGGACUGG-UUGACUGGGAAA .......(((......---.(.(((((.((((...-....)))).))))).)..(((((((.....))))))).---(((((..(((..(....)..)-)))))))))).. ( -36.70) >DroSim_CAF1 15857 103 - 1 AGAAAAGUCCGCAGGG---GGGCGGGGGAAAAGUU-UUUAUUUUACCUCGGCGAGCUCAAGUGAAACUUGAGUG---CAGUGGGGACUGGGGGACUGG-UUGACUGGGAAA .......(((.(((..---..((.((((.(((((.-...))))).)))).))(.(((((((.....))))))).---)......(((..(....)..)-))..)))))).. ( -35.40) >DroEre_CAF1 15705 88 - 1 AGAAAAGUCGGCAGGG-----GCGGGGGAAAAGUU-UUUAUUUUACCUCGGCGAGCUCAAGUGAAACUUGAGUG---CAGUGUG--------------AGGAGAUGGGAAA .......((.((....-----)).)).......((-(((((((..(((((((..(((((((.....))))))).---..)).))--------------)))))))))))). ( -23.00) >consensus AGAAAAGUCCGCAGGG_____GCGGGGGAAAAGUU_UUUAUUUUACCUCGGCGAGCUCAAGUGAAACUUGAGUG___CAGUGGGGACU___G____GG_AUAACUGGAAAA .......................((...((((........)))).)).......(((((((.....)))))))...................................... ( -5.34 = -6.12 + 0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:56 2006