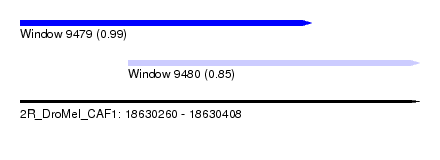
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,630,260 – 18,630,408 |
| Length | 148 |
| Max. P | 0.985700 |

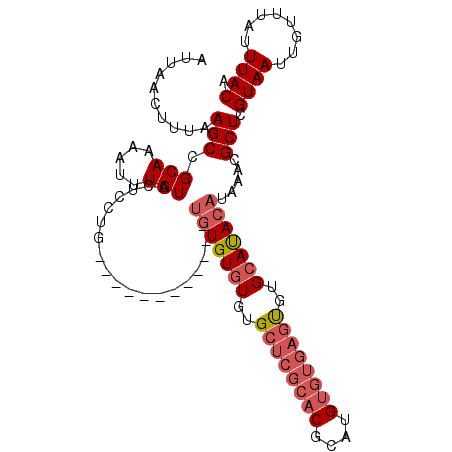
| Location | 18,630,260 – 18,630,368 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.47 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -17.79 |
| Energy contribution | -19.73 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.77 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18630260 108 + 20766785 AUUAACUUUAAGCCGCAAAAUUUGUACUCUUGUGUAUGCGAAUGUGUGUGUGUGCUCGCACGCAUGUGUGAGUGUGCAUACAUAAACGCUCGUAAUUGUUUAUUUACA .......................((((......))))(((..((((((((((..(((((((....)))))))..))))))))))..)))..((((........)))). ( -33.80) >DroSec_CAF1 40893 96 + 1 AUUAACUUUAAGCCGCAAAAUUUGUACUCCUG------------UGUGUGUGUGCUCGCACGCAUGUGUGAGUGUGCAUACAUAAACGCUCGUAAUUGUUUAUUUACA ..........(((.(((.....))).....((------------((((((((..(((((((....)))))))..))))))))))...))).((((........)))). ( -29.10) >DroSim_CAF1 37306 96 + 1 AUUAACUUUAAGCCGCAAAAUUUGUACUCCUG------------UGUGUGUGUGCUCGCACGCAUGUGUGAGUGUGCAUACAUAAACGCUCGUAAUUGUUUAUUUACA ..........(((.(((.....))).....((------------((((((((..(((((((....)))))))..))))))))))...))).((((........)))). ( -29.10) >DroAna_CAF1 45685 86 + 1 AUUAACUUUAAGCCGCAAAAUUUGUGCUCUUGUG------------UGUUUGGGCU----------UCUGCCCGGGCACACAUAAGCGCUCGUAAUUGUUUAUUUACA .......................(((((..((((------------(((((((((.----------...)))))))))))))..)))))..((((........)))). ( -33.30) >consensus AUUAACUUUAAGCCGCAAAAUUUGUACUCCUG____________UGUGUGUGUGCUCGCACGCAUGUGUGAGUGUGCAUACAUAAACGCUCGUAAUUGUUUAUUUACA ..........(((.(((.....)))...................(((((((..((((((((....))))))))..))))))).....))).((((........)))). (-17.79 = -19.73 + 1.94)

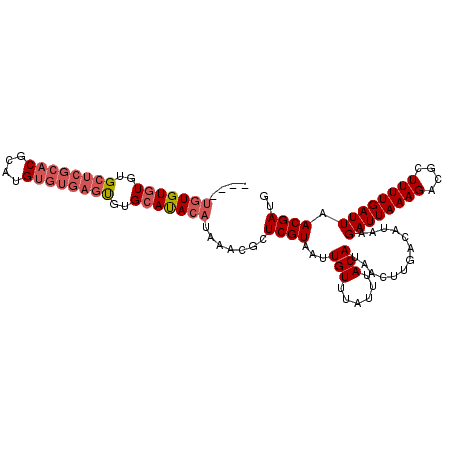
| Location | 18,630,300 – 18,630,408 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 87.74 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -19.79 |
| Energy contribution | -21.73 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18630300 108 + 20766785 AAUGUGUGUGUGUGCUCGCACGCAUGUGUGAGUGUGCAUACAUAAACGCUCGUAAUUGUUUAUUUACAUAACUUGACAUAAGAUUAAAGACGCUUUUGAUUAACGAUG ..((((((((((..(((((((....)))))))..)))))))))).....((((...(((......))).............((((((((....)))))))).)))).. ( -31.30) >DroSec_CAF1 40925 104 + 1 ----UGUGUGUGUGCUCGCACGCAUGUGUGAGUGUGCAUACAUAAACGCUCGUAAUUGUUUAUUUACAUAACAUGACAUAAGAUUAAAGACGCUUUUGAUUAACGAUG ----((((((((..(((((((....)))))))..)))))))).......((((...(((......))).............((((((((....)))))))).)))).. ( -28.40) >DroSim_CAF1 37338 104 + 1 ----UGUGUGUGUGCUCGCACGCAUGUGUGAGUGUGCAUACAUAAACGCUCGUAAUUGUUUAUUUACAUAACUUGACAUAAGAUUAAAGACGCUUUUGAUUAACGAUG ----((((((((..(((((((....)))))))..)))))))).......((((...(((......))).............((((((((....)))))))).)))).. ( -28.40) >DroAna_CAF1 45719 92 + 1 ------UGUUUGGGCU----------UCUGCCCGGGCACACAUAAGCGCUCGUAAUUGUUUAUUUACAUAACUUGACAUAAGAUUAAAGACGCUUUUGAUUAACGAUG ------(((((((((.----------...)))))))))..((.(((((.((((((........))))....((((...))))......))))))).)).......... ( -23.80) >consensus ____UGUGUGUGUGCUCGCACGCAUGUGUGAGUGUGCAUACAUAAACGCUCGUAAUUGUUUAUUUACAUAACUUGACAUAAGAUUAAAGACGCUUUUGAUUAACGAUG ....(((((((..((((((((....))))))))..))))))).......((((...(((......))).............((((((((....)))))))).)))).. (-19.79 = -21.73 + 1.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:46 2006