| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,627,776 – 18,627,869 |
| Length | 93 |
| Max. P | 0.500000 |

| Location | 18,627,776 – 18,627,869 |
|---|---|
| Length | 93 |
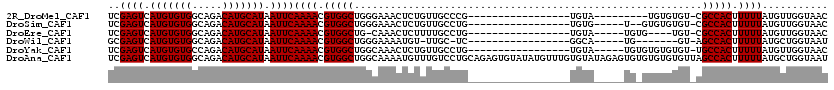
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.78 |
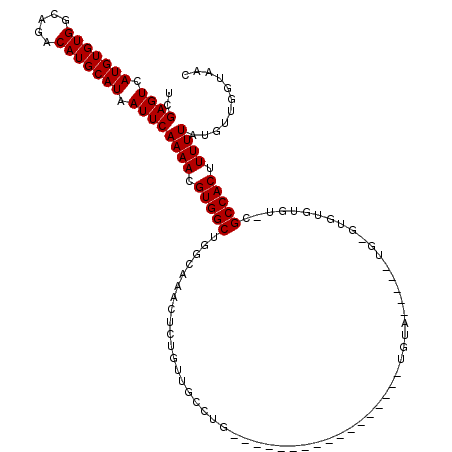
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -16.48 |
| Energy contribution | -16.48 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18627776 93 + 20766785 UCGAGUCAUGUGUGGCAGACAUGCAUAAUUCAAAACGUGGCUGGGAAACUCUGUUGCCCG-----------------UGUA---------UGUGUGU-CGCCACUUUUUAUGUUGGUAAC .(.((.((((.(((((.((((((((((.......(((.(((.((....)).....)))))-----------------).))---------)))))))-))))))....)))))).).... ( -31.20) >DroSim_CAF1 33062 95 + 1 UCGAGUCAUGUGUGGCAGACAUGCAUAAUUCAAAACGUGGCUGGGAAACUCUGUUGCCUG-----------------UGUG-----U--GUGUGUGU-CGCCACUUUUUAUGUUGGUAAC .(.((.((((.(((((.((((((((((...((....(..((.((....))..))..)...-----------------))..-----)--))))))))-))))))....)))))).).... ( -31.80) >DroEre_CAF1 40700 92 + 1 UCGAGUCAUGUGUGGCAGACAUGCAUAAUUCAAAACGUGGCUG-CAAACUCUUUUGCCUG-----------------UGUA-----UGUG----UGU-CGCCACUUUUUAUGUUGGUAAC .(.((.((((.(((((.((((((((((.......(((.(((..-...........)))))-----------------).))-----))))----)))-))))))....)))))).).... ( -26.32) >DroWil_CAF1 65895 88 + 1 GCGAGUCAUGUGUGGCAGACAUGCAUAAUUCAAAACGUGGCUGGGAAAAUGU-UUGC-UC-----------------GGCA-----UG-------GU-AGCCACUUUUUAUGCUGGUAAU ((((((.(((((((.....))))))).))))((((.(((((((.....((((-(...-..-----------------))))-----).-------.)-)))))).))))..))....... ( -25.00) >DroYak_CAF1 37363 97 + 1 UCGAGUCAUGUGUGCCAGACAUGCAUAAUUCAAAACGUGGCUGGCAAACUCUGUUGCCUG-----------------UGUA-----UGUGUGUGUGU-UGCCACUUUUUAUGUUGGUAAC ..((((......((((((.((((............)))).)))))).)))).((((((.(-----------------..((-----.(((.((....-.)))))....))..).)))))) ( -26.10) >DroAna_CAF1 43761 120 + 1 UCGAGUCAUGUGUGGCAGACAUGCAUAAUUCAAAACGUGGCUGGCAAAAUGUUUGUCCUGCAGAGUGUAUAUGUUUGUGUAUAGAGUGUGUGUGUGUUAGCCACUUUUUAUGCUGGUAAU .(.((.((((.(((((.((((((((((((((...((...((.(((((.....)))))..))...))((((((....)))))).)))).)))))))))).)))))....)))))).).... ( -36.10) >consensus UCGAGUCAUGUGUGGCAGACAUGCAUAAUUCAAAACGUGGCUGGCAAACUCUGUUGCCUG_________________UGUA_____UG_GUGUGUGU_CGCCACUUUUUAUGUUGGUAAC ..((((.(((((((.....))))))).))))((((.(((((..........................................................))))).))))........... (-16.48 = -16.48 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:44 2006