| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,627,335 – 18,627,444 |
| Length | 109 |
| Max. P | 0.888033 |

| Location | 18,627,335 – 18,627,444 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 88.81 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -19.40 |
| Energy contribution | -20.12 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18627335 109 + 20766785 GAAAAAACUAUACCAUGAAG------AUAUUUUCCAUCAGGGAAAACUCUGCUCCACACGCAGAGGUAAUUGACUUUGAAAUGAGCUCCCCUUGGCUGGAACUUCAGAAGCAUAA ...............(((((------.....(((((((((((....((((((.......))))))......(((((......))).)).)))))).))))))))))......... ( -26.10) >DroSec_CAF1 38064 108 + 1 GAA-AAACUAUGCCAUGAAG------AUAUUUUCCAUCAGGGAAAACUCUGCUCCACACGCAGAGGUAAUUGACUUUGAAAUGAGCUCCCCUUGGCUGGAACUUCAGAUGCAUAA ...-....(((((..(((((------.....(((((((((((....((((((.......))))))......(((((......))).)).)))))).))))))))))...))))). ( -31.90) >DroSim_CAF1 32626 108 + 1 GAA-AAACUACACCAUGAAG------AUAUUUUCCAUCAGGGAAAACUCUGCUCCACACGCAGAGGUAAUUGACUUUGAAAUGAGCUCCCCUUGGCUGGAACUUCAGAUGCAUAA ...-...........(((((------.....(((((((((((....((((((.......))))))......(((((......))).)).)))))).))))))))))......... ( -26.10) >DroEre_CAF1 40271 114 + 1 CGA-AAACUAUACCAUGAAUUUUCCAUCAUUUUCCAUCAGAGAAAACUCUGCCAUACAUGCAGAGGUAAUUGACUUUGAAAUGAGCUCCCCUUGGCUGGAACCUCAGAUGCAUAA ...-...........(((...((((((((((((...((((..(...((((((.......)))))).)..))))....)))))))(((......))))))))..)))......... ( -21.60) >DroYak_CAF1 36936 108 + 1 CAA-AAACUGUACCAUGAAG------ACAGUUUCCAUCAGAGAAAACUCCGCCAUACAUGAGGAGGUAAUUGACUUUGAAAUGAGCUUCCCUUGGCUGGAACCUCAGAUGCAUAA ...-(((((((.........------))))))).((((.(((.....(((((((.....(.((((((.(((........)))..))))))).)))).)))..))).))))..... ( -25.30) >consensus GAA_AAACUAUACCAUGAAG______AUAUUUUCCAUCAGGGAAAACUCUGCUCCACACGCAGAGGUAAUUGACUUUGAAAUGAGCUCCCCUUGGCUGGAACUUCAGAUGCAUAA ...............(((((...........(((((((((((....((((((.......))))))......(((((......))).)).)))))).))))))))))......... (-19.40 = -20.12 + 0.72)

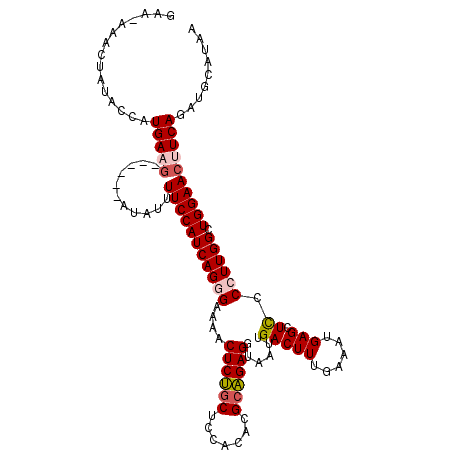
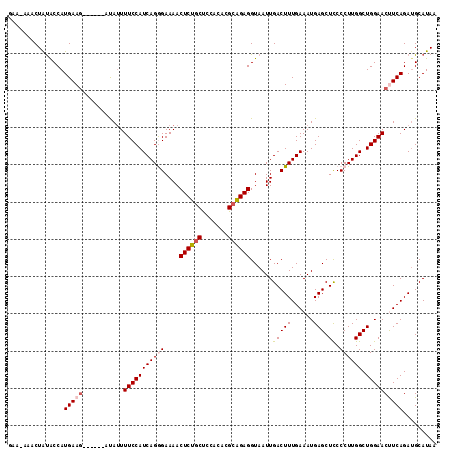
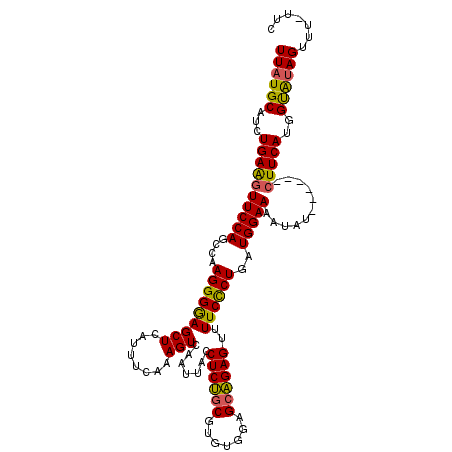
| Location | 18,627,335 – 18,627,444 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 88.81 |
| Mean single sequence MFE | -30.31 |
| Consensus MFE | -26.26 |
| Energy contribution | -25.58 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18627335 109 - 20766785 UUAUGCUUCUGAAGUUCCAGCCAAGGGGAGCUCAUUUCAAAGUCAAUUACCUCUGCGUGUGGAGCAGAGUUUUCCCUGAUGGAAAAUAU------CUUCAUGGUAUAGUUUUUUC (((((((..((((((((((....(((((((((........))).......((((((.......))))))..))))))..))))).....------))))).)))))))....... ( -30.80) >DroSec_CAF1 38064 108 - 1 UUAUGCAUCUGAAGUUCCAGCCAAGGGGAGCUCAUUUCAAAGUCAAUUACCUCUGCGUGUGGAGCAGAGUUUUCCCUGAUGGAAAAUAU------CUUCAUGGCAUAGUUU-UUC ((((((...((((((((((....(((((((((........))).......((((((.......))))))..))))))..))))).....------)))))..))))))...-... ( -32.00) >DroSim_CAF1 32626 108 - 1 UUAUGCAUCUGAAGUUCCAGCCAAGGGGAGCUCAUUUCAAAGUCAAUUACCUCUGCGUGUGGAGCAGAGUUUUCCCUGAUGGAAAAUAU------CUUCAUGGUGUAGUUU-UUC ...((((((((((((((((....(((((((((........))).......((((((.......))))))..))))))..))))).....------))))).))))))....-... ( -32.70) >DroEre_CAF1 40271 114 - 1 UUAUGCAUCUGAGGUUCCAGCCAAGGGGAGCUCAUUUCAAAGUCAAUUACCUCUGCAUGUAUGGCAGAGUUUUCUCUGAUGGAAAAUGAUGGAAAAUUCAUGGUAUAGUUU-UCG ((((((...((((.((((.......)))).))))((((...((((.....((((((.......))))))((((((.....)))))))))).)))).......))))))...-... ( -29.00) >DroYak_CAF1 36936 108 - 1 UUAUGCAUCUGAGGUUCCAGCCAAGGGAAGCUCAUUUCAAAGUCAAUUACCUCCUCAUGUAUGGCGGAGUUUUCUCUGAUGGAAACUGU------CUUCAUGGUACAGUUU-UUG .(((((((..(((((..........((((.....))))..........)))))...))))))).(((((....)))))..(((((((((------((....)).)))))))-)). ( -27.05) >consensus UUAUGCAUCUGAAGUUCCAGCCAAGGGGAGCUCAUUUCAAAGUCAAUUACCUCUGCGUGUGGAGCAGAGUUUUCCCUGAUGGAAAAUAU______CUUCAUGGUAUAGUUU_UUC ((((((...((((((((((....(((((((((........))).......((((((.......))))))..))))))..)))))...........)))))..))))))....... (-26.26 = -25.58 + -0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:43 2006