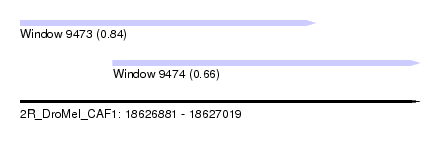
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,626,881 – 18,627,019 |
| Length | 138 |
| Max. P | 0.839171 |

| Location | 18,626,881 – 18,626,983 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.58 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -17.96 |
| Energy contribution | -18.68 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.839171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
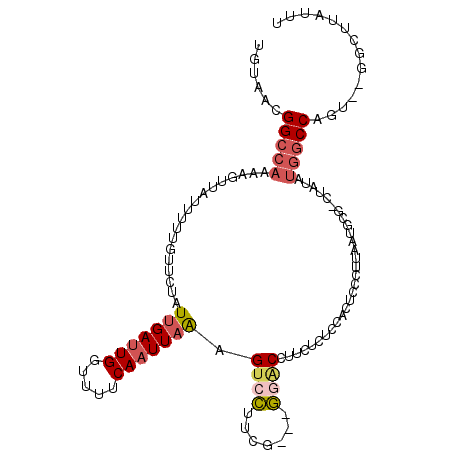
>2R_DroMel_CAF1 18626881 102 + 20766785 AUUUGCAACUUGGCCCAAUCAGUCGGACGUGAUGUAACGGCCAAAAGUUAUUUUUGUUCUAUUGAUUGGUUUUCAAUUAAAGUCCUUCG---GGACGUUCGCUUC ....(((((((((.(((((((((.(((((....(((((........)))))...))))).)))))))))...)))).....((((....---))))))).))... ( -32.10) >DroSec_CAF1 37603 102 + 1 AUUUGCAACUUGGCCCAAUCAGUCGGACGUGAUGUAACGGCCAAAAGAUAUUUUUGUUCUAUUGAUUGGUUUUCAAUUAAAGUCCUUCG---GGACGAUCUCUCC .........((((.(((((((((.(((((.(((((............)))))..))))).)))))))))...)))).....((((....---))))......... ( -25.50) >DroSim_CAF1 32165 102 + 1 AUUUGCAACUUGGCCCAAUCAGUCGGACGUGAUGUAACGGCCAAAAGUUAUUUUUGUUCUAUUGAUUGGUUUUCAAUUAAAGUCCUUCG---GGACCUUAUCUCC .........((((.(((((((((.(((((....(((((........)))))...))))).)))))))))...)))).(((.((((....---)))).)))..... ( -28.70) >DroEre_CAF1 39805 102 + 1 AUUUGCAACUUGGCCCAAUCAGUCGGACGUGAUGUAACGGCCAAAAGUUAUUUUCGUUCUAUUGAUUGGUUUUCAAUUAAAGUCCUUCG---GUGCCUUCUCUCU ...........((((((((((((.(((((....(((((........)))))...))))).)))))))))...............(....---).)))........ ( -25.70) >DroYak_CAF1 36439 102 + 1 AUUUGCAACUUGGCCCAAUCAGACGGACGUGAUGUAACGGCCAAAAGUUAUUUUUGUUCUAUUGAUUGGUUUUCAAUUAAAGUCCUUCG---GGGCUUUCUCUCU .........((((.((((((((..(((((....(((((........)))))...)))))..))))))))...))))..(((((((....---)))))))...... ( -27.40) >DroAna_CAF1 42763 96 + 1 AUUCGCAACUCGCCCCAAUCGGCUGAAUGUCACAUAAUGGG----GCCUGUCAGGGGCCUAUUGAUUGGUUGCCAUUUUGAGAUGUUUAUUUUUGCCUC-----C ....((((...(((......)))((((((((.((.((((((----(((.(((((.......))))).)))).))))).)).))))))))...))))...-----. ( -27.10) >consensus AUUUGCAACUUGGCCCAAUCAGUCGGACGUGAUGUAACGGCCAAAAGUUAUUUUUGUUCUAUUGAUUGGUUUUCAAUUAAAGUCCUUCG___GGACCUUCUCUCC ..............(((((((((.(((((....(((((........)))))...))))).)))))))))............((((.......))))......... (-17.96 = -18.68 + 0.73)



| Location | 18,626,913 – 18,627,019 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 69.21 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -12.19 |
| Energy contribution | -12.98 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18626913 106 + 20766785 UGUAACGGCCAAAAGUUAUUUUUGUUCUAUUGAUUGGUUUUCAAUUAAAGUCCUUCG---GGACGUUCGCUUCACUCCCUUAAUGUG-CUAUAUGGCCAGU--UGUUUAUUU ..(((((((((.((((.............(((((((.....))))))).((((....---))))....))))(((.........)))-.....))))).))--))....... ( -24.70) >DroSec_CAF1 37635 106 + 1 UGUAACGGCCAAAAGAUAUUUUUGUUCUAUUGAUUGGUUUUCAAUUAAAGUCCUUCG---GGACGAUCUCUCCACUCCCUUAAUACG-CUACAAGGCCAGU--UGUUUAUUU ..((((((((...................(((((((.....))))))).((((....---)))).......................-......)))).))--))....... ( -22.80) >DroSim_CAF1 32197 106 + 1 UGUAACGGCCAAAAGUUAUUUUUGUUCUAUUGAUUGGUUUUCAAUUAAAGUCCUUCG---GGACCUUAUCUCCACUCCCUUAAUGCG-CUAUAUGGCCAGU--UGUUUAUUU ..(((((((((..(((.............(((((((.....))))))).((((....---))))......................)-))...))))).))--))....... ( -22.90) >DroEre_CAF1 39837 91 + 1 UGUAACGGCCAAAAGUUAUUUUCGUUCUAUUGAUUGGUUUUCAAUUAAAGUCCUUCG---GUGCCUUCUCUCU------------------UAUGGCCGGCCUGGCCUGUUU ......(((((............(..((.(((((((.....)))))))))..)...(---(((((........------------------...)))..))))))))..... ( -19.80) >DroYak_CAF1 36471 106 + 1 UGUAACGGCCAAAAGUUAUUUUUGUUCUAUUGAUUGGUUUUCAAUUAAAGUCCUUCG---GGGCUUUCUCUCU-CUUAUUUCCUUCG-CUAUAUGGCCAGUCUGGCAUGUU- (((.(((((((..(((.............(((((((.....)))))))(((((....---)))))........-............)-))...))))).))...)))....- ( -22.80) >DroAna_CAF1 42795 98 + 1 CAUAAUGGG----GCCUGUCAGGGGCCUAUUGAUUGGUUGCCAUUUUGAGAUGUUUAUUUUUGCCUC-----CGGGCACUAAAGGUGGCUA---GGCCACG--GGCUUACUU .......((----((((((....((((((..........((((((((.((...........((((..-----..)))))).))))))))))---)))))))--))))).... ( -31.60) >consensus UGUAACGGCCAAAAGUUAUUUUUGUUCUAUUGAUUGGUUUUCAAUUAAAGUCCUUCG___GGACCUUCUCUCCACUCCCUUAAUGCG_CUAUAUGGCCAGU__GGCUUAUUU ......(((((..................(((((((.....))))))).((((.......)))).............................))))).............. (-12.19 = -12.98 + 0.79)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:41 2006