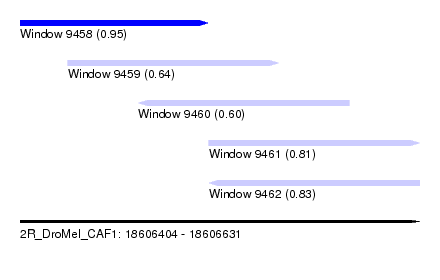
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,606,404 – 18,606,631 |
| Length | 227 |
| Max. P | 0.948013 |

| Location | 18,606,404 – 18,606,511 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 94.73 |
| Mean single sequence MFE | -30.86 |
| Consensus MFE | -28.92 |
| Energy contribution | -28.68 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18606404 107 + 20766785 C---UGCUCAUCCGCUGCACUGCUCAUCCGGAGACAAUCGUGAGCGAAAUUUAAAUGCUGCUGACUUUGCAGCUCACUGAAUGACAACUAUACGGCGAGGGGAACAAAAA .---((.((..((((((....((((((..(....)....))))))(..(((((...(((((.......)))))....)))))..).......))))).)..)).)).... ( -28.30) >DroSec_CAF1 16005 107 + 1 C---UACUCAUCCGCUGCACUGCUCAUCCGGAGACAAUCGUGAGCGAAAUUUAAAUGCUGCUGACUGAGCAGCUCUCUGAAUGACAACUAUACGGCGAGGGGAACAAAAA .---..(((...(((((...((.((((.((((((...(((....))).........((((((.....)))))))))))).))))))......)))))..)))........ ( -31.00) >DroSim_CAF1 16635 107 + 1 C---UGCUCAUCCGCUGCACUGCUCAUCCGGAGACAAUCGUGAGCGAAAUUUAAAUGCUGCUGACUUUGCAGCUCACUGAAUGACAACUAUACGGCGAGGGGAACAAAAA .---((.((..((((((....((((((..(....)....))))))(..(((((...(((((.......)))))....)))))..).......))))).)..)).)).... ( -28.30) >DroEre_CAF1 16009 106 + 1 ----UGCUCAUCCGCUGCACUGCUCAUCCGGAGACAAUCGUGAGCGAAAUUUAAAUGCUGCUGACUUUGCAGCUCACUGAAUGACAACUAUACAGCGAGGGGAAGAAAAA ----...((.(((.((((...((((((..(....)....))))))(..(((((...(((((.......)))))....)))))..).........)).)).))).)).... ( -29.00) >DroYak_CAF1 15945 110 + 1 CUGCUCCUCAUCCGCUGCACUGCUCAUCCGGAGACAAUCGUGAGCGAAAUUUAAAUGCUGCUGACUUUGCAGCUCACUGAAUGGCAAGUGUACAGCGAGGGGAACAAAAA .((.(((((...(((((((((((((((..(....)....))))))(..(((((...(((((.......)))))....)))))..).))))..))))).))))).)).... ( -37.70) >consensus C___UGCUCAUCCGCUGCACUGCUCAUCCGGAGACAAUCGUGAGCGAAAUUUAAAUGCUGCUGACUUUGCAGCUCACUGAAUGACAACUAUACGGCGAGGGGAACAAAAA ......(((...(((((....((((((..(....)....))))))(..(((((...(((((.......)))))....)))))..).......)))))..)))........ (-28.92 = -28.68 + -0.24)

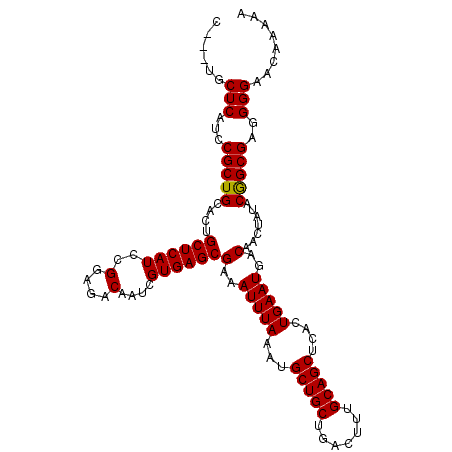
| Location | 18,606,431 – 18,606,551 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -27.71 |
| Consensus MFE | -22.74 |
| Energy contribution | -23.74 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18606431 120 + 20766785 GAGACAAUCGUGAGCGAAAUUUAAAUGCUGCUGACUUUGCAGCUCACUGAAUGACAACUAUACGGCGAGGGGAACAAAAAUAAUGGAAAGGCAACAAGCGCAGCAAAAAAUGUCGAUUGA ..((((.(((....)))........((((((...((((.((.(((.(((.(((.....))).))).)))(....)........)).))))((.....)))))))).....))))...... ( -28.80) >DroSec_CAF1 16032 120 + 1 GAGACAAUCGUGAGCGAAAUUUAAAUGCUGCUGACUGAGCAGCUCUCUGAAUGACAACUAUACGGCGAGGGGAACAAAAAUAAUGGAAAGGCAACAAGCGCAGCAAAAAAUGUCGAUUGA ....(((((.........(((((...((((((.....))))))....)))))((((........(((..(....)..............(....)...))).........))))))))). ( -24.73) >DroSim_CAF1 16662 120 + 1 GAGACAAUCGUGAGCGAAAUUUAAAUGCUGCUGACUUUGCAGCUCACUGAAUGACAACUAUACGGCGAGGGGAACAAAAAUAAUGGAAAGGCAACAAGCGCAGCAAAAAAUGUCGAUUGA ..((((.(((....)))........((((((...((((.((.(((.(((.(((.....))).))).)))(....)........)).))))((.....)))))))).....))))...... ( -28.80) >DroEre_CAF1 16035 120 + 1 GAGACAAUCGUGAGCGAAAUUUAAAUGCUGCUGACUUUGCAGCUCACUGAAUGACAACUAUACAGCGAGGGGAAGAAAAAUAAUGGAAAAGCAACAAGCGCAGCAAAAAAUCCCGAUUGA ....((((((.((............((((((...((((.(..(((.(((.(((.....))).))).)))).))))...............((.....)))))))).....)).)))))). ( -26.33) >DroYak_CAF1 15975 120 + 1 GAGACAAUCGUGAGCGAAAUUUAAAUGCUGCUGACUUUGCAGCUCACUGAAUGGCAAGUGUACAGCGAGGGGAACAAAAAUAAUGGAAAAGCAACAAGCGCAGCAAAAAAUGUCGAUUGA ..((((.(((....)))........((((((...((((((.(..((((........))))..).))))))....................((.....)))))))).....))))...... ( -29.90) >consensus GAGACAAUCGUGAGCGAAAUUUAAAUGCUGCUGACUUUGCAGCUCACUGAAUGACAACUAUACGGCGAGGGGAACAAAAAUAAUGGAAAGGCAACAAGCGCAGCAAAAAAUGUCGAUUGA ..((((.(((....)))........((((((...((((.((.(((.(((.(((.....))).))).)))(....)........)).))))((.....)))))))).....))))...... (-22.74 = -23.74 + 1.00)

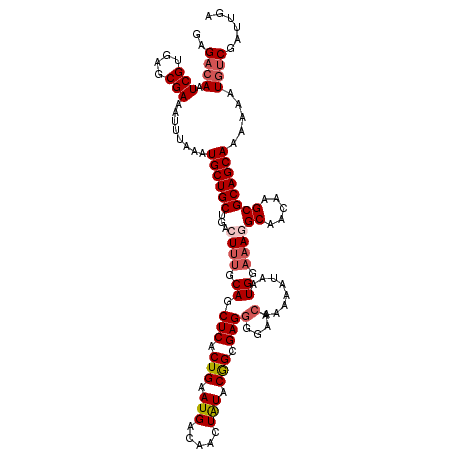
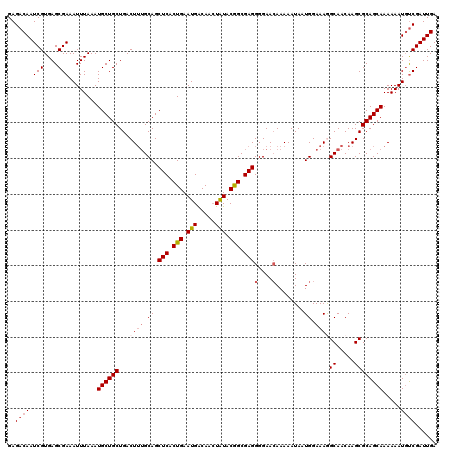
| Location | 18,606,471 – 18,606,591 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -25.45 |
| Consensus MFE | -20.34 |
| Energy contribution | -20.82 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18606471 120 - 20766785 AUUCGCUUGAGCGAAAUACCAUUUUGUUGAUUGAACAUUUUCAAUCGACAUUUUUUGCUGCGCUUGUUGCCUUUCCAUUAUUUUUGUUCCCCUCGCCGUAUAGUUGUCAUUCAGUGAGCU ....((...(((((((........(((((((((((....)))))))))))..)))))))))(((..(((.....(.(((((....((.......))...))))).).....)))..))). ( -25.30) >DroSec_CAF1 16072 120 - 1 AUUCGAUCGAGCGAAAUGCCAUUUUGUUGAUUGAACAUUUUCAAUCGACAUUUUUUGCUGCGCUUGUUGCCUUUCCAUUAUUUUUGUUCCCCUCGCCGUAUAGUUGUCAUUCAGAGAGCU ...(((.((((((....((.....(((((((((((....)))))))))))......))..)))))))))................................((((.((.....)).)))) ( -26.30) >DroSim_CAF1 16702 120 - 1 AUUCGCUCGAGCGAAAUGCCAUUUUGUUGAUUGAACAUUUUCAAUCGACAUUUUUUGCUGCGCUUGUUGCCUUUCCAUUAUUUUUGUUCCCCUCGCCGUAUAGUUGUCAUUCAGUGAGCU ....((.((((((....((.....(((((((((((....)))))))))))......))..))))))..)).....................(((((.(.............).))))).. ( -27.82) >DroEre_CAF1 16075 120 - 1 AUUUGUUUGAGCGAAAUGCCAUUUUGUUGAUUGAACAUUUUCAAUCGGGAUUUUUUGCUGCGCUUGUUGCUUUUCCAUUAUUUUUCUUCCCCUCGCUGUAUAGUUGUCAUUCAGUGAGCU ....((.((((((....((.....(.(((((((((....))))))))).)......))..))))))..)).....................(((((((.............))))))).. ( -23.72) >DroYak_CAF1 16015 120 - 1 AUUUGUUUGAGCGAAAUGCCAUUUUGCUGAUUGAACAUUUUCAAUCGACAUUUUUUGCUGCGCUUGUUGCUUUUCCAUUAUUUUUGUUCCCCUCGCUGUACACUUGCCAUUCAGUGAGCU ........((((((((.((.....((.((((((((....)))))))).))......))...((.....))...........))))))))..(((((((.............))))))).. ( -24.12) >consensus AUUCGCUUGAGCGAAAUGCCAUUUUGUUGAUUGAACAUUUUCAAUCGACAUUUUUUGCUGCGCUUGUUGCCUUUCCAUUAUUUUUGUUCCCCUCGCCGUAUAGUUGUCAUUCAGUGAGCU ....((.((((((....((.....(((((((((((....)))))))))))......))..))))))..)).....................(((((.(.............).))))).. (-20.34 = -20.82 + 0.48)

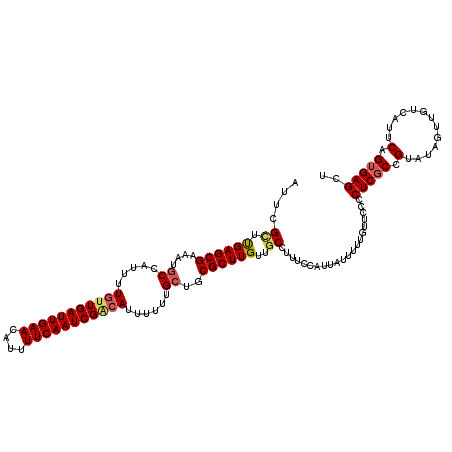
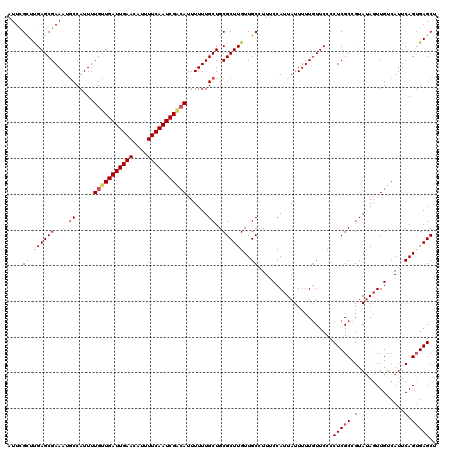
| Location | 18,606,511 – 18,606,631 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.50 |
| Mean single sequence MFE | -30.91 |
| Consensus MFE | -22.06 |
| Energy contribution | -22.58 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18606511 120 + 20766785 UAAUGGAAAGGCAACAAGCGCAGCAAAAAAUGUCGAUUGAAAAUGUUCAAUCAACAAAAUGGUAUUUCGCUCAAGCGAAUUGUUAAAGGCGCUUCCUGAAAUGUGGCGCAAGCUCAAUAU ....((((.(....)..((((.........(((.(((((((....))))))).)))...(..((.(((((....))))).))..)...)))))))).....((.(((....))))).... ( -33.10) >DroSec_CAF1 16112 114 + 1 UAAUGGAAAGGCAACAAGCGCAGCAAAAAAUGUCGAUUGAAAAUGUUCAAUCAACAAAAUGGCAUUUCGCUCGAUCGAAUUGUUGAAGGCGAUUCCUGGAAUGUGGCGCA------AUAU .........(....)..((((.........(((.(((((((....))))))).))).....((((((((...(((((..((....))..)))))..)))))))).)))).------.... ( -29.10) >DroSim_CAF1 16742 120 + 1 UAAUGGAAAGGCAACAAGCGCAGCAAAAAAUGUCGAUUGAAAAUGUUCAAUCAACAAAAUGGCAUUUCGCUCGAGCGAAUUGUUGAAGGCGAUUCCGGGAAUGUGGCGCAAGCUCAAUAU ...(((((.(....)...(((.........(((.(((((((....))))))).)))....((((.(((((....))))).))))....))).)))))....((.(((....))))).... ( -33.30) >DroEre_CAF1 16115 120 + 1 UAAUGGAAAAGCAACAAGCGCAGCAAAAAAUCCCGAUUGAAAAUGUUCAAUCAACAAAAUGGCAUUUCGCUCAAACAAAUUGUUGAAGGCGAUUCGUGAAAUGUGGCGCAGGCUUGAUAG ........((((.....((((.............(((((((....))))))).........(((((((((...(((.....)))(((.....)))))))))))).))))..))))..... ( -28.70) >DroYak_CAF1 16055 120 + 1 UAAUGGAAAAGCAACAAGCGCAGCAAAAAAUGUCGAUUGAAAAUGUUCAAUCAGCAAAAUGGCAUUUCGCUCAAACAAAUUGUCGAAGGCGAUUCUUAAAAUGUGGUGCCAGCGCGAUAG .................((((.........(((.(((((((....))))))).)))...((((((..(((.......((((((.....))))))........)))))))))))))..... ( -30.36) >consensus UAAUGGAAAGGCAACAAGCGCAGCAAAAAAUGUCGAUUGAAAAUGUUCAAUCAACAAAAUGGCAUUUCGCUCAAACGAAUUGUUGAAGGCGAUUCCUGAAAUGUGGCGCAAGCUCAAUAU .................((((.........(((.(((((((....))))))).))).....((((((((......)(((((((.....)))))))..))))))).))))........... (-22.06 = -22.58 + 0.52)

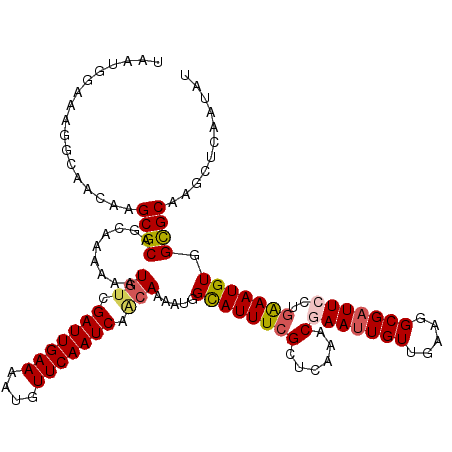
| Location | 18,606,511 – 18,606,631 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.50 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -21.46 |
| Energy contribution | -21.78 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18606511 120 - 20766785 AUAUUGAGCUUGCGCCACAUUUCAGGAAGCGCCUUUAACAAUUCGCUUGAGCGAAAUACCAUUUUGUUGAUUGAACAUUUUCAAUCGACAUUUUUUGCUGCGCUUGUUGCCUUUCCAUUA ....((.((....)))).....(((.((((((.........(((((....))))).........(((((((((((....))))))))))).........)))))).)))........... ( -30.50) >DroSec_CAF1 16112 114 - 1 AUAU------UGCGCCACAUUCCAGGAAUCGCCUUCAACAAUUCGAUCGAGCGAAAUGCCAUUUUGUUGAUUGAACAUUUUCAAUCGACAUUUUUUGCUGCGCUUGUUGCCUUUCCAUUA ....------..............((((..((..((........)).((((((....((.....(((((((((((....)))))))))))......))..))))))..))..)))).... ( -27.40) >DroSim_CAF1 16742 120 - 1 AUAUUGAGCUUGCGCCACAUUCCCGGAAUCGCCUUCAACAAUUCGCUCGAGCGAAAUGCCAUUUUGUUGAUUGAACAUUUUCAAUCGACAUUUUUUGCUGCGCUUGUUGCCUUUCCAUUA ...(((((...(((((........))...))).)))))......((.((((((....((.....(((((((((((....)))))))))))......))..))))))..)).......... ( -29.10) >DroEre_CAF1 16115 120 - 1 CUAUCAAGCCUGCGCCACAUUUCACGAAUCGCCUUCAACAAUUUGUUUGAGCGAAAUGCCAUUUUGUUGAUUGAACAUUUUCAAUCGGGAUUUUUUGCUGCGCUUGUUGCUUUUCCAUUA ....((((((.(((....((..(.....((((.((.(((.....))).))))))..............(((((((....))))))))..))....))).).))))).............. ( -23.50) >DroYak_CAF1 16055 120 - 1 CUAUCGCGCUGGCACCACAUUUUAAGAAUCGCCUUCGACAAUUUGUUUGAGCGAAAUGCCAUUUUGCUGAUUGAACAUUUUCAAUCGACAUUUUUUGCUGCGCUUGUUGCUUUUCCAUUA .....((((.((((..............((((.((.(((.....))).))))))..........((.((((((((....)))))))).)).....))))))))................. ( -25.00) >consensus AUAUCGAGCUUGCGCCACAUUUCAGGAAUCGCCUUCAACAAUUCGCUUGAGCGAAAUGCCAUUUUGUUGAUUGAACAUUUUCAAUCGACAUUUUUUGCUGCGCUUGUUGCCUUUCCAUUA ........................((((..((....(((((..(((...(((((((........(((((((((((....)))))))))))..)))))))))).)))))))..)))).... (-21.46 = -21.78 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:29 2006