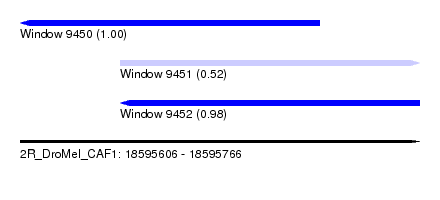
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,595,606 – 18,595,766 |
| Length | 160 |
| Max. P | 0.997163 |

| Location | 18,595,606 – 18,595,726 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.00 |
| Mean single sequence MFE | -37.17 |
| Consensus MFE | -30.01 |
| Energy contribution | -29.80 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.18 |
| Mean z-score | -4.84 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.997163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18595606 120 - 20766785 GGAAAUUUUCCGAUGAUUUGGAAGCGAUUUCUCCAGAUGACAUAGAAAUUGUUUCUAGAUCUCGCAAAAUUUCAAAUUACAACUCAAGCGAAGCGUGUCAGAACGACAUGUUCGAUUUGG .((((((((.(((.((((((((((((((((((...........))))))))))))))))))))).))))))))...........((((((((.((((((.....)))))))))).)))). ( -43.30) >DroEre_CAF1 5469 105 - 1 GGAAAUUUUCCGAUGAUUUAGAAACGAUUACUCUAGAUGCGAUAGAAAUCGUUUUUAGAUCUCGCAAAAUUUCAAAUUACAACUUGGGCGAA---------------CUGUUCAAUUUGG .((((((((.(((.(((((((((((((((..((((.......)))))))))))))))))))))).))))))))......(((.((((((...---------------..)))))).))). ( -34.40) >DroYak_CAF1 5387 120 - 1 GGAAAUUUUCCGAUGAUUUAGAAACGAUUUCUCUAGAAGCUAUAGAAAUUGUUUUUAGAUCUCGCAAAAUUUCUAAUUACAACUCAGGCGAAACCGAUCAGAGCGACCUCUUUAAUUUGA (((((((((.(((.((((((((((((((((((.(((...))).))))))))))))))))))))).)))))))))........(((.((((....)).)).)))................. ( -33.80) >consensus GGAAAUUUUCCGAUGAUUUAGAAACGAUUUCUCUAGAUGCCAUAGAAAUUGUUUUUAGAUCUCGCAAAAUUUCAAAUUACAACUCAGGCGAA_C___UCAGA_CGACCUGUUCAAUUUGG .((((((((.(((.((((((((((((((((((...........))))))))))))))))))))).))))))))......(((.(((((((..................))))))).))). (-30.01 = -29.80 + -0.21)

| Location | 18,595,646 – 18,595,766 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.11 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -17.55 |
| Energy contribution | -18.33 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18595646 120 + 20766785 GUAAUUUGAAAUUUUGCGAGAUCUAGAAACAAUUUCUAUGUCAUCUGGAGAAAUCGCUUCCAAAUCAUCGGAAAAUUUCCAAUGACAGCUGAAAUAAAAAUUAUAUGUCUGUAAUUACUA ((((((.((((((((.((((((.(((((.....))))).)))...(((((.......))))).....))).))))))))....((((..(((........)))..))))...)))))).. ( -24.90) >DroEre_CAF1 5494 118 + 1 GUAAUUUGAAAUUUUGCGAGAUCUAAAAACGAUUUCUAUCGCAUCUAGAGUAAUCGUUUCUAAAUCAUCGGAAAAUUUCCAGUGACAGCCGAAAUAAAAAUUGUAGGUC--UAACUUUUC ((.(((.((((((((.((((((.((.((((((((((((.......))))..)))))))).)).))).))).)))))))).))).)).(((..(((....)))...))).--......... ( -25.60) >DroYak_CAF1 5427 117 + 1 GUAAUUAGAAAUUUUGCGAGAUCUAAAAACAAUUUCUAUAGCUUCUAGAGAAAUCGUUUCUAAAUCAUCGGAAAAUUUCCAGUGACAGCCGAAAUAAAAAUUGUGUAUCCGUAAC---UC ((.(((.((((((((.((((((.((.((((.((((((.(((...))).)))))).)))).)).))).))).)))))))).))).)).((.((.(((.......))).)).))...---.. ( -23.00) >consensus GUAAUUUGAAAUUUUGCGAGAUCUAAAAACAAUUUCUAUAGCAUCUAGAGAAAUCGUUUCUAAAUCAUCGGAAAAUUUCCAGUGACAGCCGAAAUAAAAAUUGUAUGUC_GUAACU__UC ((.(((.((((((((.((((((.((.((((.((((((...........)))))).)))).)).))).))).)))))))).))).)).................................. (-17.55 = -18.33 + 0.78)

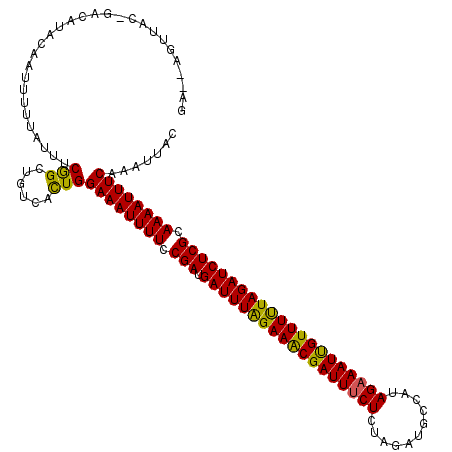
| Location | 18,595,646 – 18,595,766 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.11 |
| Mean single sequence MFE | -32.87 |
| Consensus MFE | -28.99 |
| Energy contribution | -28.00 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.19 |
| Mean z-score | -4.17 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18595646 120 - 20766785 UAGUAAUUACAGACAUAUAAUUUUUAUUUCAGCUGUCAUUGGAAAUUUUCCGAUGAUUUGGAAGCGAUUUCUCCAGAUGACAUAGAAAUUGUUUCUAGAUCUCGCAAAAUUUCAAAUUAC ..((((((...((((...(((....))).....))))....((((((((.(((.((((((((((((((((((...........))))))))))))))))))))).)))))))).)))))) ( -34.40) >DroEre_CAF1 5494 118 - 1 GAAAAGUUA--GACCUACAAUUUUUAUUUCGGCUGUCACUGGAAAUUUUCCGAUGAUUUAGAAACGAUUACUCUAGAUGCGAUAGAAAUCGUUUUUAGAUCUCGCAAAAUUUCAAAUUAC .(((((((.--.......)))))))....(((......)))((((((((.(((.(((((((((((((((..((((.......)))))))))))))))))))))).))))))))....... ( -29.80) >DroYak_CAF1 5427 117 - 1 GA---GUUACGGAUACACAAUUUUUAUUUCGGCUGUCACUGGAAAUUUUCCGAUGAUUUAGAAACGAUUUCUCUAGAAGCUAUAGAAAUUGUUUUUAGAUCUCGCAAAAUUUCUAAUUAC .(---((.((((...(..(((....)))..).)))).)))(((((((((.(((.((((((((((((((((((.(((...))).))))))))))))))))))))).)))))))))...... ( -34.40) >consensus GA__AGUUAC_GACAUACAAUUUUUAUUUCGGCUGUCACUGGAAAUUUUCCGAUGAUUUAGAAACGAUUUCUCUAGAUGCCAUAGAAAUUGUUUUUAGAUCUCGCAAAAUUUCAAAUUAC .............................(((......)))((((((((.(((.((((((((((((((((((...........))))))))))))))))))))).))))))))....... (-28.99 = -28.00 + -0.99)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:19 2006