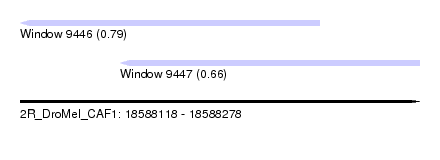
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,588,118 – 18,588,278 |
| Length | 160 |
| Max. P | 0.785263 |

| Location | 18,588,118 – 18,588,238 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.58 |
| Mean single sequence MFE | -34.78 |
| Consensus MFE | -28.76 |
| Energy contribution | -27.92 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18588118 120 - 20766785 UUUUCCACCCAUUGUUUCGUAUGACGUCAUGCCGAUGAACACGGCGUAUACGUUAUACGCUUGACCUAUCCGCUUCAUUGUUGGUAACCGAAUAGACCGUCUUUGAAGGGUUUCGCAGUC ..................(((((((((.((((((.......))))))..)))))))))((..((((......(((((..(.((((..........)))).)..)))))))))..)).... ( -33.00) >DroSec_CAF1 6345 120 - 1 UUUUCCACCCAUUGCUUCGUAUGACGUCAUGCCGAUGAACACGGCGUAUACGUUAUACGCUUGACCUAUCCGCUUCAUUGUUGGUAACCGAAUGGACCGCAUUUGAAGGGUUUCGCAGUG .........((((((..((((((((((.((((((.......))))))..))))))))))..........((.(((((.(((.(((..((....))))))))..)))))))....)))))) ( -38.30) >DroSim_CAF1 6332 120 - 1 UUUUCCACCCAUUGCUUCGUAUGACGUCAUGCCGAUGAACACGGCGUAUACGUUAUACGCUUGACCUAUCCGCUUCAUUGUUGGUAACCGAAUGGACCGCAUUUGAAGGGUUUCGCAGUG .........((((((..((((((((((.((((((.......))))))..))))))))))..........((.(((((.(((.(((..((....))))))))..)))))))....)))))) ( -38.30) >DroEre_CAF1 6275 120 - 1 CUUUCCACCGGUUGCUUCAUAUGACGUCAUGCCGAUGAACACGGCGUAUACGUUAUAUGCUUGACCUAUCCACUUCAUUGUUGGUAACCGACUAAGCCGCCUUUGAUGUGUCUCGUAGUA .........(((((...((((((((((.((((((.......))))))..))))))))))..)))))....(((.(((..(.((((..........)))).)..))).))).......... ( -31.70) >DroYak_CAF1 6401 120 - 1 UUUUCCACCAGUUACUUUGUAUGACGUCAUGCCGAUGCACACGGCGUAUACGUUAUACGCUUCACCUAUCCGCCUCAUUGCUGGUAACCGAAUAAACCGCUUGCGAUGGGUUUCGUAGUA ..................(((((((((.((((((.......))))))..))))))))).......((((..(((.((((((((((..........))))...)))))))))...)))).. ( -32.60) >consensus UUUUCCACCCAUUGCUUCGUAUGACGUCAUGCCGAUGAACACGGCGUAUACGUUAUACGCUUGACCUAUCCGCUUCAUUGUUGGUAACCGAAUAGACCGCAUUUGAAGGGUUUCGCAGUA ..........(((((..((((((((((.((((((.......))))))..))))))))))...........(.(((((....((((..........))))....))))).)....))))). (-28.76 = -27.92 + -0.84)

| Location | 18,588,158 – 18,588,278 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -23.69 |
| Energy contribution | -23.37 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18588158 120 - 20766785 UAAGCAAGCAAUAUAUCCGAGUAAUCCGCGACUGCCUGAAUUUUCCACCCAUUGUUUCGUAUGACGUCAUGCCGAUGAACACGGCGUAUACGUUAUACGCUUGACCUAUCCGCUUCAUUG ((((((((((((......((....)).((....))...............))))))).(((((((((.((((((.......))))))..))))))))))))))................. ( -28.60) >DroSec_CAF1 6385 120 - 1 UAAGCAAGCAAUAUAUCCGAGUAAUCCGCGACUGCCUGAAUUUUCCACCCAUUGCUUCGUAUGACGUCAUGCCGAUGAACACGGCGUAUACGUUAUACGCUUGACCUAUCCGCUUCAUUG ((((((((((((......((....)).((....))...............))))))).(((((((((.((((((.......))))))..))))))))))))))................. ( -31.00) >DroSim_CAF1 6372 120 - 1 UAAGCAAGCAAUAUAUCCGAGUAAUCCGCGACUGCCUGAAUUUUCCACCCAUUGCUUCGUAUGACGUCAUGCCGAUGAACACGGCGUAUACGUUAUACGCUUGACCUAUCCGCUUCAUUG ((((((((((((......((....)).((....))...............))))))).(((((((((.((((((.......))))))..))))))))))))))................. ( -31.00) >DroEre_CAF1 6315 120 - 1 UAACCAAACAAUAUAUCUGAGUAAGCCUCGACUGCUUGAACUUUCCACCGGUUGCUUCAUAUGACGUCAUGCCGAUGAACACGGCGUAUACGUUAUAUGCUUGACCUAUCCACUUCAUUG .................(((((((((.......)))))...........(((((...((((((((((.((((((.......))))))..))))))))))..))))).......))))... ( -26.80) >DroYak_CAF1 6441 120 - 1 UAACCAAGCAAUAUAUCCGAGUAGUCCGCGACUGCUUGAAUUUUCCACCAGUUACUUUGUAUGACGUCAUGCCGAUGCACACGGCGUAUACGUUAUACGCUUCACCUAUCCGCCUCAUUG .....((((........(((((((((...)))))))))....................(((((((((.((((((.......))))))..))))))))))))).................. ( -33.20) >consensus UAAGCAAGCAAUAUAUCCGAGUAAUCCGCGACUGCCUGAAUUUUCCACCCAUUGCUUCGUAUGACGUCAUGCCGAUGAACACGGCGUAUACGUUAUACGCUUGACCUAUCCGCUUCAUUG ....(((((.........(((((((.........................))))))).(((((((((.((((((.......))))))..))))))))))))))................. (-23.69 = -23.37 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:14 2006