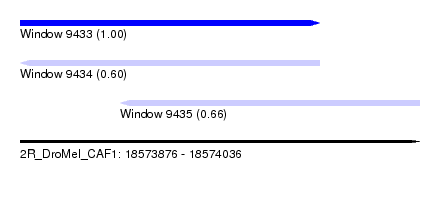
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,573,876 – 18,574,036 |
| Length | 160 |
| Max. P | 0.999612 |

| Location | 18,573,876 – 18,573,996 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.08 |
| Mean single sequence MFE | -51.02 |
| Consensus MFE | -47.96 |
| Energy contribution | -48.20 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.78 |
| SVM RNA-class probability | 0.999612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18573876 120 + 20766785 AGCCAGGGUCAGGUCAACAGGCUGACCACCUUCUGGCAGGAGUCCGAUGUGGACAUGUCCCGUGGCCUUGAUUUUGUGCCGCGCGGCCAUGUCCUCGCUCGGUUCACCCAUCUGCAGCAC .((((((((..(((((......))))))))..)))))..(((.((((((.(((((((.(((((((((........).)))))).)).))))))).)).)))))))............... ( -52.60) >DroSec_CAF1 53590 120 + 1 AGCCAGGGUCAGGUUAACAAGCUGACCACCUUCUGGCAGGAGUCCGAUGUGGACAUGUCCCGUGGCAUUGACUUCGUGCCGCGCGGCCAUGUCCUCGCUCGGUUCACCCAUCUGCAGCAC .(((((((((((.(.....).)))))).....)))))..(((.((((((.(((((((.((((((((((.......)))))))).)).))))))).)).)))))))............... ( -53.70) >DroSim_CAF1 49532 120 + 1 AGCCAGGGUCAGGUGAACAAGCUGACCACCUUCUGGCAGGAGUCCGAUGUGGACAUGUCCCGUGGCCUUGACUUCGUGCCGCGCGGCCAUGUCCUCGCUCGGUUCACUCAUCUGCAGCAC .(((((((((((.(.....).)))))).....)))))..((((((((((.(((((((.((((((((..((....)).)))))).)).))))))).)).))))...))))........... ( -54.20) >DroEre_CAF1 55697 120 + 1 AGCCAGGGUCAGGUCAACAGGCUGACCACCUUCUGGCAGGAGUCCGAUGUGGACAUGUCCCGCGGCCUUGAUUUUGUGCCACGCGGCCAUGUCCUCGCUCGGUUCACCCAUCUGCAGCAC .((((((((..(((((......))))))))..)))))..(((.((((((.(((((((.((((.((((........).))).)).)).))))))).)).)))))))............... ( -49.10) >DroYak_CAF1 55425 120 + 1 AGCCAGGGUAAGAUCAACAGGCUGACCACAUUCUGGCAGGAGUCCGAUGUGGACAUGUCCCGCGGCCUUGAUUUCGUGCCGCGCGGCCACGUCCUCGCUCGGUUCACCCAUCUGCAGCAC .((((((((..(.(((......))).)..))))))))..(((.((((((.((((.((.((((((((..((....)).)))))).)).)).)))).)).)))))))............... ( -45.50) >consensus AGCCAGGGUCAGGUCAACAGGCUGACCACCUUCUGGCAGGAGUCCGAUGUGGACAUGUCCCGUGGCCUUGAUUUCGUGCCGCGCGGCCAUGUCCUCGCUCGGUUCACCCAUCUGCAGCAC .(((((((((((.(.....).)))))).....)))))..(((.((((((.(((((((.((((((((...........)))))).)).))))))).)).)))))))............... (-47.96 = -48.20 + 0.24)


| Location | 18,573,876 – 18,573,996 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.08 |
| Mean single sequence MFE | -48.86 |
| Consensus MFE | -43.76 |
| Energy contribution | -43.88 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18573876 120 - 20766785 GUGCUGCAGAUGGGUGAACCGAGCGAGGACAUGGCCGCGCGGCACAAAAUCAAGGCCACGGGACAUGUCCACAUCGGACUCCUGCCAGAAGGUGGUCAGCCUGUUGACCUGACCCUGGCU ...........(((....((((..(.(((((((.((.((.(((.(........)))).)))).))))))).).))))...)))(((((..(((((((((....))))))..)))))))). ( -48.60) >DroSec_CAF1 53590 120 - 1 GUGCUGCAGAUGGGUGAACCGAGCGAGGACAUGGCCGCGCGGCACGAAGUCAAUGCCACGGGACAUGUCCACAUCGGACUCCUGCCAGAAGGUGGUCAGCUUGUUAACCUGACCCUGGCU ..(((((((..((((...((((..(.(((((((.((.((.((((.........)))).)))).))))))).).)))))))))))).....((.((((((.........))))))))))). ( -45.90) >DroSim_CAF1 49532 120 - 1 GUGCUGCAGAUGAGUGAACCGAGCGAGGACAUGGCCGCGCGGCACGAAGUCAAGGCCACGGGACAUGUCCACAUCGGACUCCUGCCAGAAGGUGGUCAGCUUGUUCACCUGACCCUGGCU ..(((((((..((((...((((..(.(((((((.((.((.(((.(........)))).)))).))))))).).)))))))))))).....((.((((((.........))))))))))). ( -47.10) >DroEre_CAF1 55697 120 - 1 GUGCUGCAGAUGGGUGAACCGAGCGAGGACAUGGCCGCGUGGCACAAAAUCAAGGCCGCGGGACAUGUCCACAUCGGACUCCUGCCAGAAGGUGGUCAGCCUGUUGACCUGACCCUGGCU ...........(((....((((..(.(((((((.((.((((((.(........))))))))).))))))).).))))...)))(((((..(((((((((....))))))..)))))))). ( -52.10) >DroYak_CAF1 55425 120 - 1 GUGCUGCAGAUGGGUGAACCGAGCGAGGACGUGGCCGCGCGGCACGAAAUCAAGGCCGCGGGACAUGUCCACAUCGGACUCCUGCCAGAAUGUGGUCAGCCUGUUGAUCUUACCCUGGCU ...........(((....((((..(.(((((((.((.((((((.(........))))))))).))))))).).))))...)))(((((..((.((((((....)))))).))..))))). ( -50.60) >consensus GUGCUGCAGAUGGGUGAACCGAGCGAGGACAUGGCCGCGCGGCACGAAAUCAAGGCCACGGGACAUGUCCACAUCGGACUCCUGCCAGAAGGUGGUCAGCCUGUUGACCUGACCCUGGCU ...........((((...((((..(.(((((((.((.((.(((...........))).)))).))))))).).))))))))..(((((..(((((((((....))))))..)))))))). (-43.76 = -43.88 + 0.12)


| Location | 18,573,916 – 18,574,036 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -48.00 |
| Consensus MFE | -44.44 |
| Energy contribution | -44.48 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18573916 120 - 20766785 CGCUGGAGUUCUCCACCUUGAUGGUGUAGCUGAAUUCGUGGUGCUGCAGAUGGGUGAACCGAGCGAGGACAUGGCCGCGCGGCACAAAAUCAAGGCCACGGGACAUGUCCACAUCGGACU ((((.(.((((.(((((.....)))(((((............))))).....)).))))).)))).(((((((.((.((.(((.(........)))).)))).))))))).......... ( -46.50) >DroSec_CAF1 53630 120 - 1 CGCUGGAGUUCUCCACCUUGAUGGUGUAGCUGAAUGCGUGGUGCUGCAGAUGGGUGAACCGAGCGAGGACAUGGCCGCGCGGCACGAAGUCAAUGCCACGGGACAUGUCCACAUCGGACU ((((.(.((((.(((((.....)))(((((............))))).....)).))))).)))).(((((((.((.((.((((.........)))).)))).))))))).......... ( -46.90) >DroSim_CAF1 49572 120 - 1 CGCUGGAGUUCUCCACCUUGAUGGUGUAGCUGAAUGCGUGGUGCUGCAGAUGAGUGAACCGAGCGAGGACAUGGCCGCGCGGCACGAAGUCAAGGCCACGGGACAUGUCCACAUCGGACU ((((.(.((((..((((.....))))..(((.(.((((......))))..).)))))))).)))).(((((((.((.((.(((.(........)))).)))).))))))).......... ( -44.40) >DroEre_CAF1 55737 120 - 1 CGCUGGAGUUCUCCACCUCGAUGGUGUAGCUGAAUGGGUGGUGCUGCAGAUGGGUGAACCGAGCGAGGACAUGGCCGCGUGGCACAAAAUCAAGGCCGCGGGACAUGUCCACAUCGGACU ((((.(.((((.(((((.....)))(((((............))))).....)).))))).)))).(((((((.((.((((((.(........))))))))).))))))).......... ( -50.00) >DroYak_CAF1 55465 120 - 1 CGCUGGAGUUCUCCACCUUGAUGGUGUAGCUGAAUGGGUGGUGCUGCAGAUGGGUGAACCGAGCGAGGACGUGGCCGCGCGGCACGAAAUCAAGGCCGCGGGACAUGUCCACAUCGGACU ((((.(.((((.(((((.....)))(((((............))))).....)).))))).)))).(((((((.((.((((((.(........))))))))).))))))).......... ( -52.20) >consensus CGCUGGAGUUCUCCACCUUGAUGGUGUAGCUGAAUGCGUGGUGCUGCAGAUGGGUGAACCGAGCGAGGACAUGGCCGCGCGGCACGAAAUCAAGGCCACGGGACAUGUCCACAUCGGACU ((((.(.((((.(((((.....)))(((((............))))).....)).))))).)))).(((((((.((.((.(((...........))).)))).))))))).......... (-44.44 = -44.48 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:57:03 2006