
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,560,824 – 18,560,987 |
| Length | 163 |
| Max. P | 0.990963 |

| Location | 18,560,824 – 18,560,916 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -23.77 |
| Consensus MFE | -13.98 |
| Energy contribution | -15.57 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18560824 92 + 20766785 AC------UAACCCAUCCAAUAUACCCU-UUACAAAUGGUUGGGUGGCGAAAAAUAAUGCAUGCCACCGCUGAUGGAUACGAGCAUCGAUGUCGAUGCU ..------....(((((........((.-........))...(((((((............)))))))...))))).....(((((((....))))))) ( -26.70) >DroSec_CAF1 40634 92 + 1 AC------UAACCCAUCCUAUAUACCCU-UUAUAAAUGGUUGGGUGGCGAAAAAUGAUGCAUGCCACCGCUGAUGGAUACGAGCAUCGAUGUCGAUGCU ..------....(((((........((.-........))...(((((((............)))))))...))))).....(((((((....))))))) ( -26.70) >DroSim_CAF1 36525 92 + 1 AC------UAACCCAUCCUAUAUACCCU-UUAUAAAUGGAUGGGUGGCGAAAAAUGAUGCAUGCCACCGCUGAUGGAUACGAGCAUCGAUGUCGAUGCU .(------((((((((((..((((....-.))))...))))))))((((....(((...))).....))))..))).....(((((((....))))))) ( -28.10) >DroEre_CAF1 42601 98 + 1 UUUAAUCAAUCCAUAAUCAAUAUACCCG-UAAUAAACGGUUCAGUGGCGAAAAAUGAUGCAUGCCACCGCUGAUGGAUACGAACAUCGAUGUCGAUGCU ........(((((............(((-(.....)))).(((((((.(....(((...)))..).)))))))))))).....(((((....))))).. ( -23.70) >DroYak_CAF1 42308 98 + 1 UAGGAUGAAAACCUUUCCAGUAUACCCG-CAAUAAAUGGUUGGGUGGCGAAAAAUGAUGCAUGCCACCGCUGAUGGAUACGAACAUCGAUGUCGAUGCU ..(((..........))).((((.((..-((((.....))))(((((((............)))))))......))))))...(((((....))))).. ( -23.60) >DroAna_CAF1 38607 94 + 1 UUU-----GAAACCUUCGUAUAUACCCCACAAGAUGUGAAACAUAGGCGAAUAAUUAUUUAUUCCACUGUUGAUGGAUACAAACAUCACUGUCGAUCUA ...-----.......................((((.(((...((((..((((((....))))))..))))(((((........)))))...))))))). ( -13.80) >consensus AC______UAACCCAUCCAAUAUACCCU_UAAUAAAUGGUUGGGUGGCGAAAAAUGAUGCAUGCCACCGCUGAUGGAUACGAACAUCGAUGUCGAUGCU ............(((((........((..........))...(((((((............)))))))...))))).....(((((((....))))))) (-13.98 = -15.57 + 1.59)

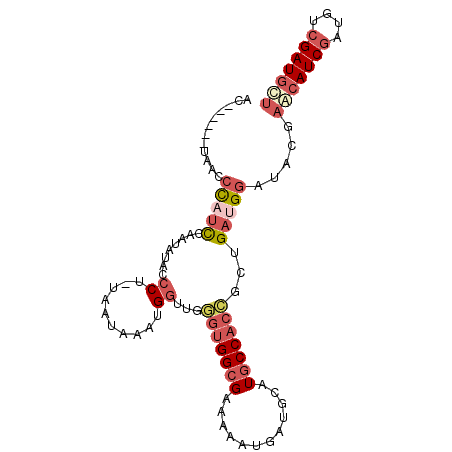
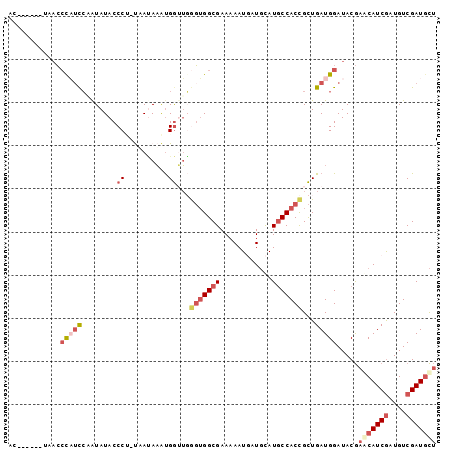
| Location | 18,560,824 – 18,560,916 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -17.15 |
| Energy contribution | -18.46 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18560824 92 - 20766785 AGCAUCGACAUCGAUGCUCGUAUCCAUCAGCGGUGGCAUGCAUUAUUUUUCGCCACCCAACCAUUUGUAA-AGGGUAUAUUGGAUGGGUUA------GU (((((((....)))))))...(((((((.(.((((((..............))))))).(((........-..)))......)))))))..------.. ( -29.54) >DroSec_CAF1 40634 92 - 1 AGCAUCGACAUCGAUGCUCGUAUCCAUCAGCGGUGGCAUGCAUCAUUUUUCGCCACCCAACCAUUUAUAA-AGGGUAUAUAGGAUGGGUUA------GU (((((((....)))))))...(((((((.(.((((((..............))))))).(((........-..)))......)))))))..------.. ( -29.54) >DroSim_CAF1 36525 92 - 1 AGCAUCGACAUCGAUGCUCGUAUCCAUCAGCGGUGGCAUGCAUCAUUUUUCGCCACCCAUCCAUUUAUAA-AGGGUAUAUAGGAUGGGUUA------GU (((((((....))))))).((((((((.....)))).)))).............((((((((...((((.-....))))..))))))))..------.. ( -31.40) >DroEre_CAF1 42601 98 - 1 AGCAUCGACAUCGAUGUUCGUAUCCAUCAGCGGUGGCAUGCAUCAUUUUUCGCCACUGAACCGUUUAUUA-CGGGUAUAUUGAUUAUGGAUUGAUUAAA .((((((....))))))(((.((((((((((((((((..............)))))))..((((.....)-))).....))))...))))))))..... ( -29.34) >DroYak_CAF1 42308 98 - 1 AGCAUCGACAUCGAUGUUCGUAUCCAUCAGCGGUGGCAUGCAUCAUUUUUCGCCACCCAACCAUUUAUUG-CGGGUAUACUGGAAAGGUUUUCAUCCUA (((((((....))))))).((((((....(.((((((..............)))))))...((.....))-.)))))).......(((.......))). ( -24.14) >DroAna_CAF1 38607 94 - 1 UAGAUCGACAGUGAUGUUUGUAUCCAUCAACAGUGGAAUAAAUAAUUAUUCGCCUAUGUUUCACAUCUUGUGGGGUAUAUACGAAGGUUUC-----AAA .(((((........(((((((.(((((.....))))))))))))....((((..((((..((((.....))))..))))..))))))))).-----... ( -21.20) >consensus AGCAUCGACAUCGAUGCUCGUAUCCAUCAGCGGUGGCAUGCAUCAUUUUUCGCCACCCAACCAUUUAUAA_AGGGUAUAUAGGAUGGGUUA______GA (((((((....)))))))...(((((((...((((((..............))))))...((..........))........))))))).......... (-17.15 = -18.46 + 1.31)

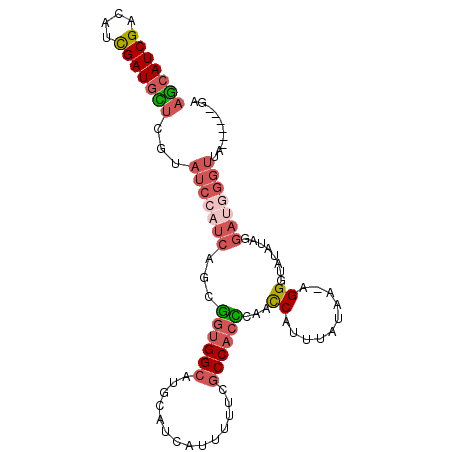
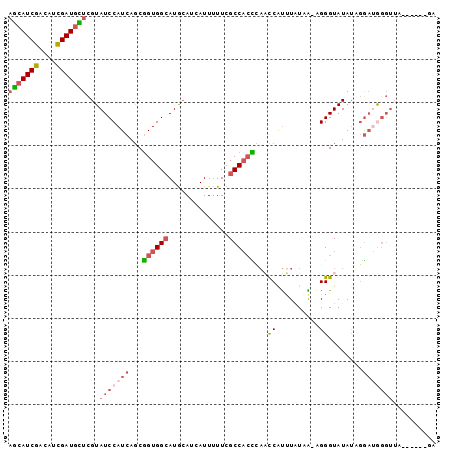
| Location | 18,560,852 – 18,560,949 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 90.24 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -25.42 |
| Energy contribution | -25.42 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18560852 97 - 20766785 CGUGAAAAGCGCGGCGUGGAAAACUCUCGUUCCAGCAUCGACAUCGAUGCUCGUAUCCAUCAGCGGUGGCAUGCAUUAUUUUUCGCCACCCAACCAU .(((.....)))(((((((((........)))))((((((....))))))..((((((((.....)))).)))).........)))).......... ( -29.70) >DroSec_CAF1 40662 97 - 1 CGUGAAAAGCGCGGCGUGGAAAACUCUCGUUCCAGCAUCGACAUCGAUGCUCGUAUCCAUCAGCGGUGGCAUGCAUCAUUUUUCGCCACCCAACCAU .(((.....)))(((((((((........)))))((((((....))))))..((((((((.....)))).)))).........)))).......... ( -29.70) >DroSim_CAF1 36553 97 - 1 CGUGAAAAGCGCGGCGUGGAAAACUCUCGUUCCAGCAUCGACAUCGAUGCUCGUAUCCAUCAGCGGUGGCAUGCAUCAUUUUUCGCCACCCAUCCAU .(((.....)))(((((((((........)))))((((((....))))))..((((((((.....)))).)))).........)))).......... ( -29.70) >DroEre_CAF1 42635 97 - 1 CGUGAAAAGCGCGGCGUGGAAAACUCUCGUUCCAGCAUCGACAUCGAUGUUCGUAUCCAUCAGCGGUGGCAUGCAUCAUUUUUCGCCACUGAACCGU .(((.....)))(((((((((........)))))((((((....))))))..((((((((.....)))).)))).........)))).......... ( -27.30) >DroYak_CAF1 42342 97 - 1 CGUGAAAAGCGCGGCGUGGAAAACUCUCGUUCCAGCAUCGACAUCGAUGUUCGUAUCCAUCAGCGGUGGCAUGCAUCAUUUUUCGCCACCCAACCAU .(((.....)))(((((((((........)))))((((((....))))))..((((((((.....)))).)))).........)))).......... ( -27.30) >DroAna_CAF1 38637 97 - 1 CGUGAAAAGCGCGGCGUGGAAAACUCUUGCUCCUAGAUCGACAGUGAUGUUUGUAUCCAUCAACAGUGGAAUAAAUAAUUAUUCGCCUAUGUUUCAC .((((((.....(((((.((....(((.......))))).))((((((((((((.(((((.....))))))))))).)))))).)))....)))))) ( -21.80) >consensus CGUGAAAAGCGCGGCGUGGAAAACUCUCGUUCCAGCAUCGACAUCGAUGCUCGUAUCCAUCAGCGGUGGCAUGCAUCAUUUUUCGCCACCCAACCAU .(((.....)))(((((((((........)))))((((((....))))))..((((((((.....)))).)))).........)))).......... (-25.42 = -25.42 + 0.00)

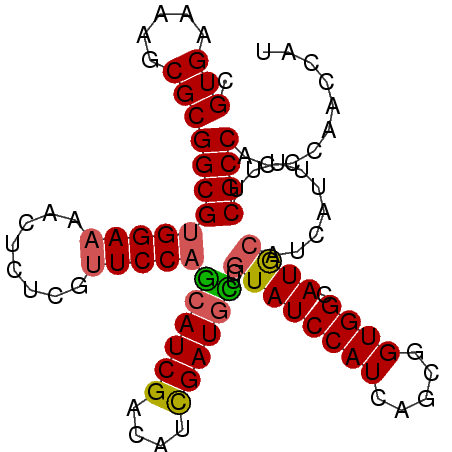
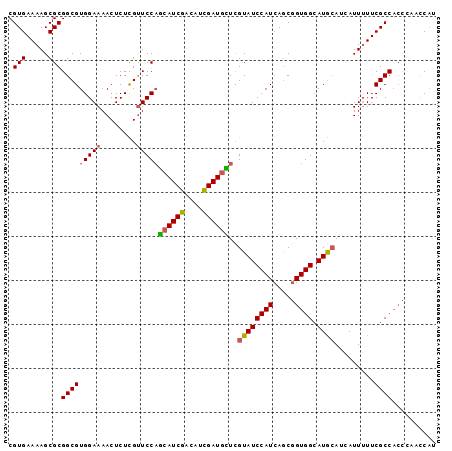
| Location | 18,560,886 – 18,560,987 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 85.61 |
| Mean single sequence MFE | -33.05 |
| Consensus MFE | -27.09 |
| Energy contribution | -27.65 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18560886 101 + 20766785 CUGAUGGAUACGAGCAUCGAUGUCGAUGCUGGAACGAGAGUUUUCCACGCCGCGCUUUUCACGGCCGCAUCCAUCAAAUCCAUGAAAUGCGCAGAGCGCGG .((((((((.(((((((((....)))))))(((((....))..)))..((((.(.....).)))))).))))))))...........(((((...))))). ( -38.60) >DroSec_CAF1 40696 94 + 1 CUGAUGGAUACGAGCAUCGAUGUCGAUGCUGGAACGAGAGUUUUCCACGCCGCGCUUUUCACGGCCGCAUCCAUCAAAUCCAUGAAAUGCGCA-------G .((((((((.(((((((((....)))))))(((((....))..)))..((((.(.....).)))))).)))))))).................-------. ( -33.10) >DroSim_CAF1 36587 101 + 1 CUGAUGGAUACGAGCAUCGAUGUCGAUGCUGGAACGAGAGUUUUCCACGCCGCGCUUUUCACGGCCGCAUCCAUCAAAUCCAUGAAAUGCGCAGAGCGCGG .((((((((.(((((((((....)))))))(((((....))..)))..((((.(.....).)))))).))))))))...........(((((...))))). ( -38.60) >DroEre_CAF1 42669 92 + 1 CUGAUGGAUACGAACAUCGAUGUCGAUGCUGGAACGAGAGUUUUCCACGCCGCGCUUUUCACGGCCGCAUCCAUCAAAUCC---------GCAGAGCGCGG .((((((((.((..(((((....))))).((((((....))..)))).((((.(.....).)))))).))))))))...((---------((.....)))) ( -33.50) >DroYak_CAF1 42376 92 + 1 CUGAUGGAUACGAACAUCGAUGUCGAUGCUGGAACGAGAGUUUUCCACGCCGCGCUUUUCACGGCCGCAUCCAUCAAAUCC---------GCAGAGCGCGG .((((((((.((..(((((....))))).((((((....))..)))).((((.(.....).)))))).))))))))...((---------((.....)))) ( -33.50) >DroAna_CAF1 38671 95 + 1 UUGAUGGAUACAAACAUCACUGUCGAUCUAGGAGCAAGAGUUUUCCACGCCGCGCUUUUCACGGUCGCAUCCAUCAA-UCCGCCGUAGCCGUA-----GGG ((((((((((((........)))(((((.((((((..(.((.......)))..))))))...))))).)))))))))-.((..((....))..-----)). ( -21.00) >consensus CUGAUGGAUACGAACAUCGAUGUCGAUGCUGGAACGAGAGUUUUCCACGCCGCGCUUUUCACGGCCGCAUCCAUCAAAUCCAUGAAAUGCGCAGAGCGCGG .((((((((.(((((((((....)))))))(((((....))..)))..((((.(.....).)))))).))))))))......................... (-27.09 = -27.65 + 0.56)

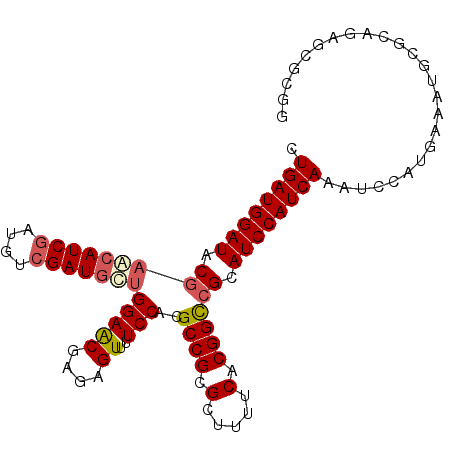
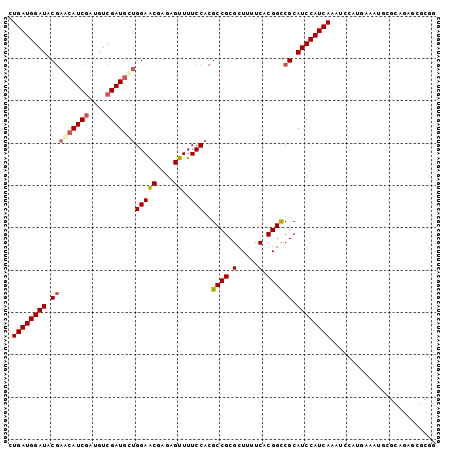
| Location | 18,560,886 – 18,560,987 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 85.61 |
| Mean single sequence MFE | -42.15 |
| Consensus MFE | -36.48 |
| Energy contribution | -36.48 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.79 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18560886 101 - 20766785 CCGCGCUCUGCGCAUUUCAUGGAUUUGAUGGAUGCGGCCGUGAAAAGCGCGGCGUGGAAAACUCUCGUUCCAGCAUCGACAUCGAUGCUCGUAUCCAUCAG ..((((...))))............(((((((((((((((((.....)))))).(((((........)))))((((((....)))))).))))))))))). ( -46.40) >DroSec_CAF1 40696 94 - 1 C-------UGCGCAUUUCAUGGAUUUGAUGGAUGCGGCCGUGAAAAGCGCGGCGUGGAAAACUCUCGUUCCAGCAUCGACAUCGAUGCUCGUAUCCAUCAG .-------.................(((((((((((((((((.....)))))).(((((........)))))((((((....)))))).))))))))))). ( -40.90) >DroSim_CAF1 36587 101 - 1 CCGCGCUCUGCGCAUUUCAUGGAUUUGAUGGAUGCGGCCGUGAAAAGCGCGGCGUGGAAAACUCUCGUUCCAGCAUCGACAUCGAUGCUCGUAUCCAUCAG ..((((...))))............(((((((((((((((((.....)))))).(((((........)))))((((((....)))))).))))))))))). ( -46.40) >DroEre_CAF1 42669 92 - 1 CCGCGCUCUGC---------GGAUUUGAUGGAUGCGGCCGUGAAAAGCGCGGCGUGGAAAACUCUCGUUCCAGCAUCGACAUCGAUGUUCGUAUCCAUCAG ((((.....))---------))...(((((((((((((((((.....)))))).(((((........)))))((((((....)))))).))))))))))). ( -44.20) >DroYak_CAF1 42376 92 - 1 CCGCGCUCUGC---------GGAUUUGAUGGAUGCGGCCGUGAAAAGCGCGGCGUGGAAAACUCUCGUUCCAGCAUCGACAUCGAUGUUCGUAUCCAUCAG ((((.....))---------))...(((((((((((((((((.....)))))).(((((........)))))((((((....)))))).))))))))))). ( -44.20) >DroAna_CAF1 38671 95 - 1 CCC-----UACGGCUACGGCGGA-UUGAUGGAUGCGACCGUGAAAAGCGCGGCGUGGAAAACUCUUGCUCCUAGAUCGACAGUGAUGUUUGUAUCCAUCAA ...-----..(.((....)).).-((((((((((((.(((((.....))))))..(((..........)))......((((....)))).))))))))))) ( -30.80) >consensus CCGCGCUCUGCGCAUUUCAUGGAUUUGAUGGAUGCGGCCGUGAAAAGCGCGGCGUGGAAAACUCUCGUUCCAGCAUCGACAUCGAUGCUCGUAUCCAUCAG .........................(((((((((((((((((.....)))))).(((((........)))))((((((....)))))).))))))))))). (-36.48 = -36.48 + 0.00)

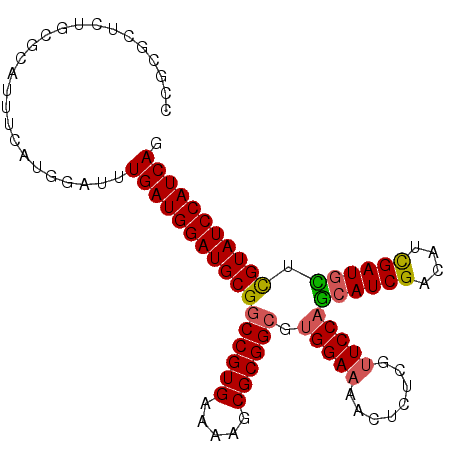
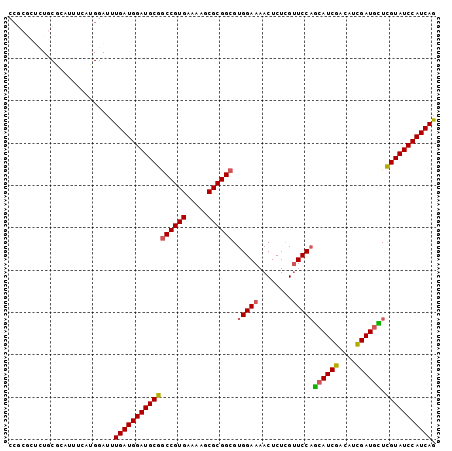
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:56 2006