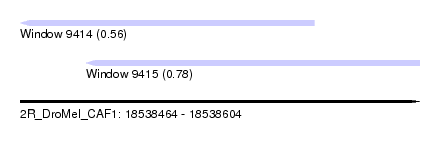
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,538,464 – 18,538,604 |
| Length | 140 |
| Max. P | 0.781188 |

| Location | 18,538,464 – 18,538,567 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 85.40 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -23.70 |
| Energy contribution | -24.42 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18538464 103 - 20766785 UCAUUUCAUUGGAGGCAUAAAGCAAGAUUUGUGUAAAUGUCAGGUGGUAAUAGUGACACUGUGCGCCACUUGAUUGGUUUUACUU------UGGCCAAGAGCC------ACAGGA-- ......((((....(((((((......))))))).))))(((((((((.((((.....))))..))))))))).........((.------((((.....)))------).))..-- ( -29.50) >DroPse_CAF1 21029 105 - 1 UCAUUUCAUUUGAGGCAUAAAGCAAGAUUUGUGUAAAUGUCAGGUGGUAAUAGUGACACUGUGCGCCACUUGAUUGGUUUUACUU------UGGCCAAGAGCC------ACACAUCC ..............((.....))..(((.((((((((.((((((((((.((((.....))))..))))))))))....)))))..------.(((.....)))------))).))). ( -28.40) >DroWil_CAF1 25543 82 - 1 UCAUUUCGUU-GAGGCAUAAGGCAAGAUUUGUGUAAAUGUCAGGUGGUAAUAGUGACACUGUGGGCCACUUGAUUGGUUUCU--U-------------------------------- ......((((-...(((((((......))))))).))))(((((((((.((((.....))))..))))))))).........--.-------------------------------- ( -23.00) >DroYak_CAF1 19392 103 - 1 UCAUUUCAUUGGAGGCAUAAAGCAAGAUUUGUGUAAAUGUCAGGUGGUAAUAGUGACACUGUGCGCCACUUGAUUGGUUUUACUU------UGGCCAAGAGCC------ACAGGA-- ......((((....(((((((......))))))).))))(((((((((.((((.....))))..))))))))).........((.------((((.....)))------).))..-- ( -29.50) >DroAna_CAF1 16570 115 - 1 UCAUUUCAUUUGAGGCAUAAAGCAAGAUUUGUGUAAAUGUCAGGUGGUAAUAGUGACACUGUGCGCCACUUGAUUGGUUUCGCUUUGGUUUUGGCCAAGAGCCACUGCCACAGGA-- ........((((.((((.(((((.(((((.........((((((((((.((((.....))))..)))))))))).))))).)))))((((((.....))))))..)))).)))).-- ( -35.90) >DroPer_CAF1 20053 105 - 1 UCAUUUCAUUUGAGGCAUAAAGCAAGAUUUGUGUAAAUGUCAGGUGGUAAUAGUGACACUGUGCGCCACUUGAUUGGUUUUACUU------UGGCCAAGAGCC------ACACAUCC ..............((.....))..(((.((((((((.((((((((((.((((.....))))..))))))))))....)))))..------.(((.....)))------))).))). ( -28.40) >consensus UCAUUUCAUUUGAGGCAUAAAGCAAGAUUUGUGUAAAUGUCAGGUGGUAAUAGUGACACUGUGCGCCACUUGAUUGGUUUUACUU______UGGCCAAGAGCC______ACAGGA__ ......((((....(((((((......))))))).))))(((((((((.((((.....))))..)))))))))((((((.............))))))................... (-23.70 = -24.42 + 0.72)

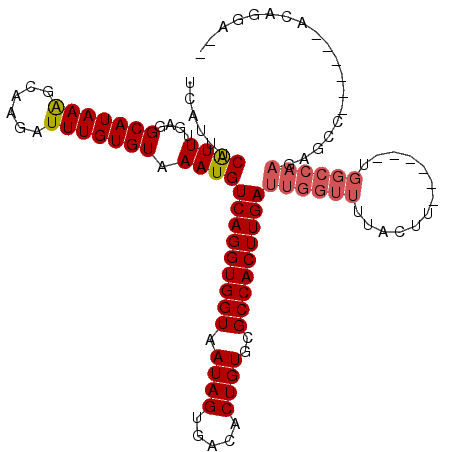
| Location | 18,538,487 – 18,538,604 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.09 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -23.36 |
| Energy contribution | -23.08 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
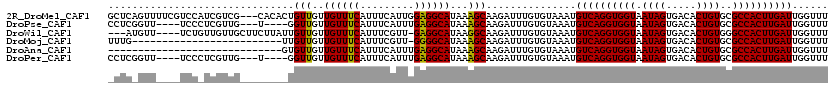
>2R_DroMel_CAF1 18538487 117 - 20766785 GCUCAGUUUUCGUCCAUCGUCG---CACACUGUUGUUGUUUCAUUUCAUUGGAGGCAUAAAGCAAGAUUUGUGUAAAUGUCAGGUGGUAAUAGUGACACUGUGCGCCACUUGAUUGGUUU .............((((((.((---((((.(((..(((((((((((((((....(((((((......))))))).))))..)))))).)))))..))).)))))).....))).)))... ( -30.80) >DroPse_CAF1 21054 109 - 1 CCUCGGUU----UCCCUCGUUG---U----GGUUGUUGUUUCAUUUCAUUUGAGGCAUAAAGCAAGAUUUGUGUAAAUGUCAGGUGGUAAUAGUGACACUGUGCGCCACUUGAUUGGUUU .(((((..----..))..(..(---(----((........))))..)....)))(((((((......)))))))....((((((((((.((((.....))))..))))))))))...... ( -27.90) >DroWil_CAF1 25546 112 - 1 ---AUGUU----UCUGUUGUUGCUUCUUAUUGUUGUUGUUUCAUUUCGUU-GAGGCAUAAGGCAAGAUUUGUGUAAAUGUCAGGUGGUAAUAGUGACACUGUGGGCCACUUGAUUGGUUU ---.....----.......((((......(((((..(((((((......)-))))))...))))).......))))..((((((((((.((((.....))))..))))))))))...... ( -30.12) >DroMoj_CAF1 24047 94 - 1 UUUG-------------------------UUGUUGUUGUUUCAUUUCGUU-GGGGCAUAAAGCAAGAUUUGUGUAAAUGUCAGGUGGUAAUAGUGACACUGUGCGCCACUUGAUUGGUUU ..(.-------------------------(((((..(((..((......)-)..)))...))))).)...........((((((((((.((((.....))))..))))))))))...... ( -28.00) >DroAna_CAF1 16605 91 - 1 -----------------------------GUGUUGUUGUUUCAUUUCAUUUGAGGCAUAAAGCAAGAUUUGUGUAAAUGUCAGGUGGUAAUAGUGACACUGUGCGCCACUUGAUUGGUUU -----------------------------.((((..(((((((.......)))))))...))))..............((((((((((.((((.....))))..))))))))))...... ( -26.40) >DroPer_CAF1 20078 109 - 1 CCUCGGUU----UCCCUCGUUG---U----GGUUGUUGUUUCAUUUCAUUUGAGGCAUAAAGCAAGAUUUGUGUAAAUGUCAGGUGGUAAUAGUGACACUGUGCGCCACUUGAUUGGUUU .(((((..----..))..(..(---(----((........))))..)....)))(((((((......)))))))....((((((((((.((((.....))))..))))))))))...... ( -27.90) >consensus _CUC_GUU____UCC_UCGUUG________UGUUGUUGUUUCAUUUCAUUUGAGGCAUAAAGCAAGAUUUGUGUAAAUGUCAGGUGGUAAUAGUGACACUGUGCGCCACUUGAUUGGUUU ...............................(((..((((((.........))))))...)))...............((((((((((.((((.....))))..))))))))))...... (-23.36 = -23.08 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:43 2006