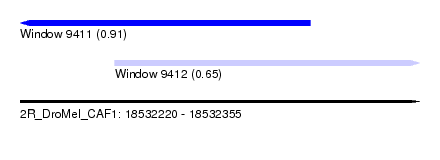
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,532,220 – 18,532,355 |
| Length | 135 |
| Max. P | 0.913067 |

| Location | 18,532,220 – 18,532,318 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.46 |
| Mean single sequence MFE | -21.96 |
| Consensus MFE | -14.76 |
| Energy contribution | -14.98 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
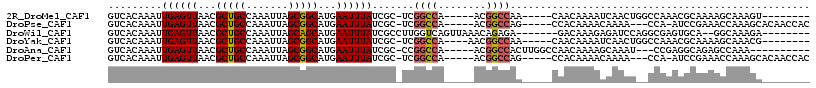
>2R_DroMel_CAF1 18532220 98 - 20766785 GUCACAAAUUGAGUUAACGCUGCCAAAUUAGCGGCAUGAAUUUAUCGC-UCGGCCA-----ACGGCCAA-----CAACAAAAUCAACUGGCCAAACGCAAAAGCAAAGU-------- .........((((((...(((((.......)))))...))))))..((-(..((..-----..(((((.-----.............)))))....))...))).....-------- ( -21.74) >DroPse_CAF1 13962 102 - 1 GUCACAAAUUGAGUUAACGCUGCCAAAUUAGCGGCAUGAAUUUAUCGC-UCGGCCA-----ACGGCCAG-----CCACAAAACAAAA---CCA-AUCCGAAACCAAAGCACAACCAC .......((((.(((...(((((.......)))))...........((-(.((((.-----..))))))-----)..........))---)))-))..................... ( -19.10) >DroWil_CAF1 17310 100 - 1 GUCACAAAUUGAGUUAACGCUGCCAAAUUAGCAGCAUGAAUUUAUCGCCUUGGUCAGUUAAACAGAGA-------GACAAAGAGAUCCAGGCGAGUGCA--GGCAAAGA-------- (((......((((((...(((((.......)))))...))))))((((((.((((.............-------........)))).)))))).....--))).....-------- ( -25.60) >DroYak_CAF1 12909 99 - 1 GUCACAAAUUGAGUUAACGCUGCCAAAUUAGCGGCAUGAAUUUAUCGC-UCGGCCA----AACGGCCAA-----CAACAAAAUCAACUGGCCAAACGCAAAAGCAAACG-------- ((((......((((....(((((.......)))))..((.....))))-))((((.----...))))..-----.............)))).....((....)).....-------- ( -21.80) >DroAna_CAF1 11457 98 - 1 GUCACAAAUUGAGUUAACGCUGCCAAAUUAGCGGCAUGAAUUUAUCGC-CCGGCCA-----ACGGCCACUUGGCCAACAAAAGCAAAU---CCGAGGCAGAGCCAAA---------- ........(((.(((...(((((.......)))))...........((-((((...-----..((((....))))..(....).....---))).)))..)))))).---------- ( -24.40) >DroPer_CAF1 12989 102 - 1 GUCACAAAUUGAGUUAACGCUGCCAAAUUAGCGGCAUGAAUUUAUCGC-UCGGCCA-----ACGGCCAG-----CCACAAAACAAAA---CCA-AUCCGAAACCAAAGCACAACCAC .......((((.(((...(((((.......)))))...........((-(.((((.-----..))))))-----)..........))---)))-))..................... ( -19.10) >consensus GUCACAAAUUGAGUUAACGCUGCCAAAUUAGCGGCAUGAAUUUAUCGC_UCGGCCA_____ACGGCCAA_____CAACAAAACCAAAU__CCAAAUGCAAAACCAAAGC________ .........((((((...(((((.......)))))...)))))).......((((........)))).................................................. (-14.76 = -14.98 + 0.22)
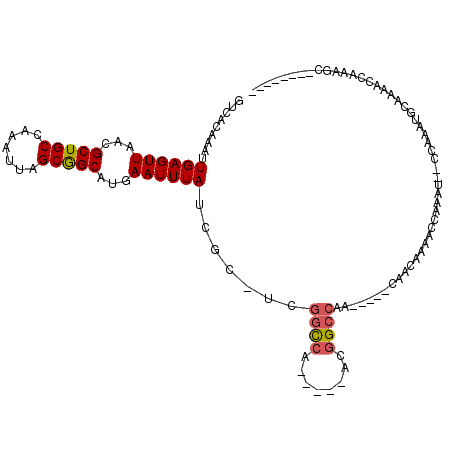
| Location | 18,532,252 – 18,532,355 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.70 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -19.50 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18532252 103 + 20766785 UUG-----UUGGCCGU-----UGG--CCG-A-GCGAUAAAUUCAUGCCGCUAAUUUGGCAGCGUUAACUCAAUUUGUGACCACAAAA-UGCACAAUUUGUCGCUGAUUUUGAUUGCCC-- ...-----((((((..-----.))--)))-)-((((((((((..(((((......)))))(((((........((((....))))))-)))..))))))))))...............-- ( -28.30) >DroVir_CAF1 12180 102 + 1 CUC------CAUUCGU-----UU----GG-A-GCGAUAAAUUUAUGCCGCUAAUUUGGCAGCGUUAACUCAAUUUGUGACCACAAAA-UGCACAAUUUGUCGCGGCUUUUGAUUGCCACA .((------((.....-----.)----))-)-((((((((((..(((((......)))))(((((........((((....))))))-)))..))))))))))(((........)))... ( -30.30) >DroGri_CAF1 13236 101 + 1 CUG------CAUUCGU-----UU----AGCG-GCGAUAAAUUUAUGCGGCUAAUUUGGCAGCGUUAACUCAAUUUGUGACCACAAAA-UGCACAAUUUGUCGCGGCUUUUGAUUGCCA-- ..(------((.(((.-----..----(((.-((((((((((.((((.(((.....))).)))).....((.(((((....))))).-))...)))))))))).)))..))).)))..-- ( -31.80) >DroWil_CAF1 17340 108 + 1 UC-------UCUCUGUUUAACUGA--CCA-AGGCGAUAAAUUCAUGCUGCUAAUUUGGCAGCGUUAACUCAAUUUGUGACCACAAAACUGAACAAUUUGUCGCUGAUUUUGAUUGCCU-- ..-------...............--...-.(((((((((((.((((((((.....))))))))....(((.(((((....)))))..)))..)))))))))))..............-- ( -25.90) >DroMoj_CAF1 16292 106 + 1 GUC------CAUUCGU-----UGGUUCAG-A-GCGAUAAAUUUAUGCCGCUAAUUUGGCAGCGUUAACUCAAUUUGUGACCACAAAG-UGCACAAUUUGUCGCGGCUCUUGAUUGCCACA ...------.....((-----.(((((((-(-((..........(((((......)))))(((......(((.(((((((......)-).))))).))).))).)))).)))..))))). ( -27.80) >DroAna_CAF1 11484 108 + 1 UUGGCCAAGUGGCCGU-----UGG--CCG-G-GCGAUAAAUUCAUGCCGCUAAUUUGGCAGCGUUAACUCAAUUUGUGACCACAAAA-UGCACAAUUUGUCGCUGAUUUUGAUUGCCC-- ..((((((.......)-----)))--))(-(-((((((((.((((((((......)))))(((......(((.(((((.(.......-.)))))).))).)))))).))).)))))))-- ( -34.70) >consensus UUG______CAUCCGU_____UGG__CCG_A_GCGAUAAAUUCAUGCCGCUAAUUUGGCAGCGUUAACUCAAUUUGUGACCACAAAA_UGCACAAUUUGUCGCGGAUUUUGAUUGCCA__ ................................((((((((((.((((.(((.....))).))))........(((((....))))).......))))))))))................. (-19.50 = -19.50 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:40 2006