
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,518,242 – 18,518,396 |
| Length | 154 |
| Max. P | 0.999838 |

| Location | 18,518,242 – 18,518,341 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 93.60 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -24.98 |
| Energy contribution | -25.18 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
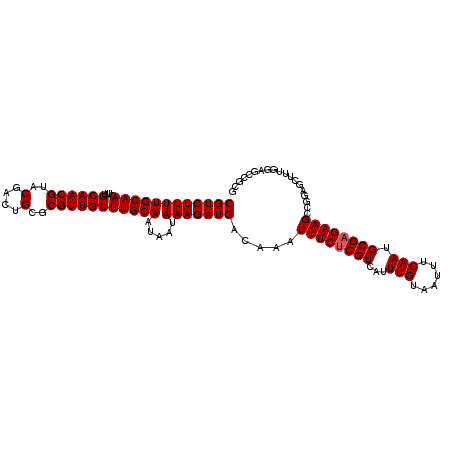
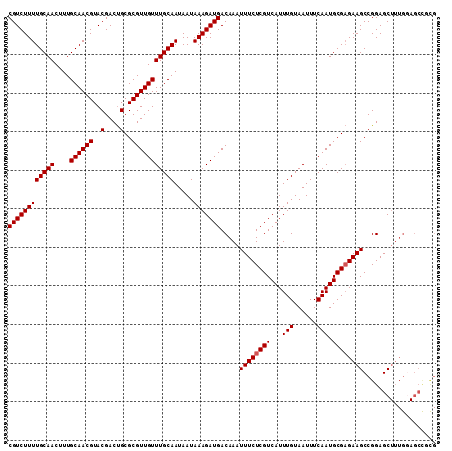
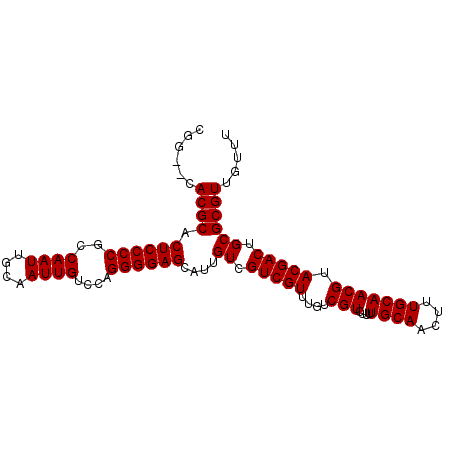
>2R_DroMel_CAF1 18518242 99 - 20766785 CGUCUUUUGCAACUUUGCAACGUACGACUGCGCGUUGUUUGCAAUAAUAAAGAUGACAAAUUUCUCGUCAUUUGUAAUUUCAAUGCGAGAAGCCGGAGC------------ ((((((((((((....((((((..(....)..))))))))))).....))))))).....((((((((...(((......))).)))))))).......------------ ( -25.90) >DroSec_CAF1 17244 111 - 1 CGUCUUUUGCAACUUUGCAACGUACGACUGCGCGUUGUUUGCAAUAAUAAAGAUGACAAAUUUCUCGUCAUUUGUAAUUUCAAUGCGAGAAGCCGGAGCUUUGGAGCCGCG ((((((((((((....((((((..(....)..))))))))))).....)))))))......(((((((...(((......))).)))))))((.((..(....)..)))). ( -30.20) >DroSim_CAF1 15661 111 - 1 CGUCUUUUGCAACUUUGCAACGUACGACUGCGCGUUGUUUGCAAUAAUAAAGAUGACAAAUUUCUCGUCAUUUGUAAUUUCAAUGCGAGAAGCCGGAGCUUUGGAGCCGCG ((((((((((((....((((((..(....)..))))))))))).....)))))))......(((((((...(((......))).)))))))((.((..(....)..)))). ( -30.20) >DroEre_CAF1 20485 111 - 1 CGUCUUUUGCAACUUUGCAACGUACGACUGCGCGUUGUUUGCAAUAAUAAAGAUGACAAAUUUCUCGUCAUUUGUAAUUUCAAUGCGAGAAGCCGGAGCUUUGGAGCUCCG ((((((((((((....((((((..(....)..))))))))))).....))))))).....((((((((...(((......))).)))))))).(((((((....))))))) ( -36.80) >DroYak_CAF1 23179 111 - 1 CGUCUUUUGCAACUUUGCAACGUACGACCGCGCGUUGUUUGCAAUAAUAAAGAUGACAAAUUUCUCGUCAUUUGUAAUUUCAAUGCGCGAAGCCGGAGCUUUGCAGCUUUG (((.(.(((((....))))).).)))..(((((((((.(((((........((((((.........)))))))))))...)))))))))....(((((((....))))))) ( -32.90) >consensus CGUCUUUUGCAACUUUGCAACGUACGACUGCGCGUUGUUUGCAAUAAUAAAGAUGACAAAUUUCUCGUCAUUUGUAAUUUCAAUGCGAGAAGCCGGAGCUUUGGAGCCGCG ((((((((((((....((((((..(....)..))))))))))).....))))))).....((((((((...(((......))).))))))))................... (-24.98 = -25.18 + 0.20)

| Location | 18,518,301 – 18,518,396 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 98.75 |
| Mean single sequence MFE | -35.14 |
| Consensus MFE | -34.64 |
| Energy contribution | -34.64 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.21 |
| SVM RNA-class probability | 0.999838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
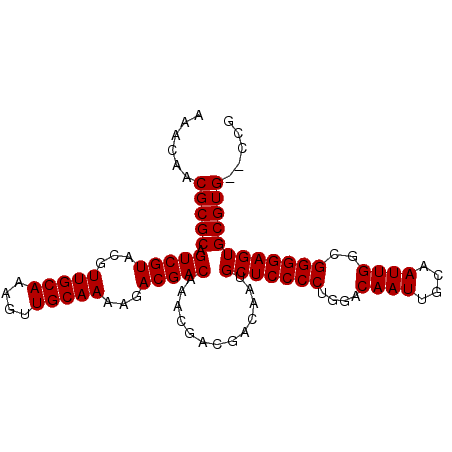
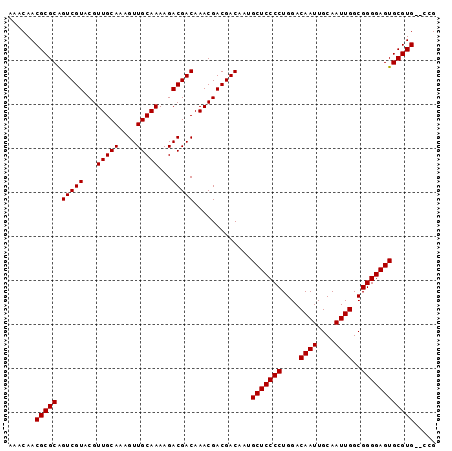
>2R_DroMel_CAF1 18518301 95 + 20766785 AAACAACGCGCAGUCGUACGUUGCAAAGUUGCAAAAGACGACAAACGACGACAAUGCUCCCCUGGACAAUUGCAAUUGGCGGGGAGUGCGUG--CCG ......(((((.(((((...(((((....)))))...))))).............(((((((....((((....))))..))))))))))))--... ( -35.00) >DroSec_CAF1 17315 95 + 1 AAACAACGCGCAGUCGUACGUUGCAAAGUUGCAAAAGACGACAAACGACGACAAUGCUCCCCUGGACAAUUGCAAUUGGCGGGGAGUGCGUG--CCG ......(((((.(((((...(((((....)))))...))))).............(((((((....((((....))))..))))))))))))--... ( -35.00) >DroSim_CAF1 15732 95 + 1 AAACAACGCGCAGUCGUACGUUGCAAAGUUGCAAAAGACGACAAACGACGACAAUGCUCCCCUGGACAAUUGCAAUUGGCGGGGAGUGCGUG--CCG ......(((((.(((((...(((((....)))))...))))).............(((((((....((((....))))..))))))))))))--... ( -35.00) >DroEre_CAF1 20556 97 + 1 AAACAACGCGCAGUCGUACGUUGCAAAGUUGCAAAAGACGACAAACGACGACAAUGCUCCCCUGGACAAUUGCAAUUGGCGGGGAGUGCGUGCGCCG .......((((.(((((...(((((....)))))...))))).....(((.....(((((((....((((....))))..))))))).))))))).. ( -35.90) >DroYak_CAF1 23250 95 + 1 AAACAACGCGCGGUCGUACGUUGCAAAGUUGCAAAAGACGACAAACGACGACAAUGCUCCCCUGGACAAUUGCAAUUGGCGGGGAGUGCGUG--CCG ......(((((.(((((...(((((....)))))...))))).............(((((((....((((....))))..))))))))))))--... ( -34.80) >consensus AAACAACGCGCAGUCGUACGUUGCAAAGUUGCAAAAGACGACAAACGACGACAAUGCUCCCCUGGACAAUUGCAAUUGGCGGGGAGUGCGUG__CCG ......(((((.(((((...(((((....)))))...))))).............(((((((....((((....))))..))))))))))))..... (-34.64 = -34.64 + 0.00)

| Location | 18,518,301 – 18,518,396 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 98.75 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -29.50 |
| Energy contribution | -29.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18518301 95 - 20766785 CGG--CACGCACUCCCCGCCAAUUGCAAUUGUCCAGGGGAGCAUUGUCGUCGUUUGUCGUCUUUUGCAACUUUGCAACGUACGACUGCGCGUUGUUU (((--(.((((((((((..((((....))))....)))))).......(((((....(((....((((....))))))).))))))))).))))... ( -31.40) >DroSec_CAF1 17315 95 - 1 CGG--CACGCACUCCCCGCCAAUUGCAAUUGUCCAGGGGAGCAUUGUCGUCGUUUGUCGUCUUUUGCAACUUUGCAACGUACGACUGCGCGUUGUUU (((--(.((((((((((..((((....))))....)))))).......(((((....(((....((((....))))))).))))))))).))))... ( -31.40) >DroSim_CAF1 15732 95 - 1 CGG--CACGCACUCCCCGCCAAUUGCAAUUGUCCAGGGGAGCAUUGUCGUCGUUUGUCGUCUUUUGCAACUUUGCAACGUACGACUGCGCGUUGUUU (((--(.((((((((((..((((....))))....)))))).......(((((....(((....((((....))))))).))))))))).))))... ( -31.40) >DroEre_CAF1 20556 97 - 1 CGGCGCACGCACUCCCCGCCAAUUGCAAUUGUCCAGGGGAGCAUUGUCGUCGUUUGUCGUCUUUUGCAACUUUGCAACGUACGACUGCGCGUUGUUU (((((((((((((((((..((((....))))....))))))...)).))).((..(((((.(.(((((....))))).).))))).))))))))... ( -33.50) >DroYak_CAF1 23250 95 - 1 CGG--CACGCACUCCCCGCCAAUUGCAAUUGUCCAGGGGAGCAUUGUCGUCGUUUGUCGUCUUUUGCAACUUUGCAACGUACGACCGCGCGUUGUUU (((--(.(((.((((((..((((....))))....)))))).......(((((....(((....((((....))))))).))))).))).))))... ( -30.80) >consensus CGG__CACGCACUCCCCGCCAAUUGCAAUUGUCCAGGGGAGCAUUGUCGUCGUUUGUCGUCUUUUGCAACUUUGCAACGUACGACUGCGCGUUGUUU ......((((.((((((..((((....))))....))))))....((.(((((....(((....((((....))))))).))))).))))))..... (-29.50 = -29.50 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:35 2006