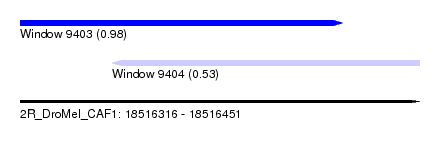
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,516,316 – 18,516,451 |
| Length | 135 |
| Max. P | 0.980763 |

| Location | 18,516,316 – 18,516,425 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -16.98 |
| Energy contribution | -17.82 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
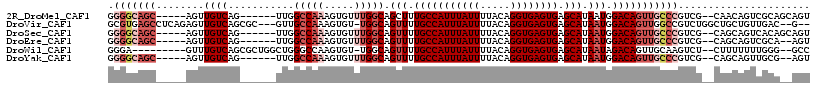
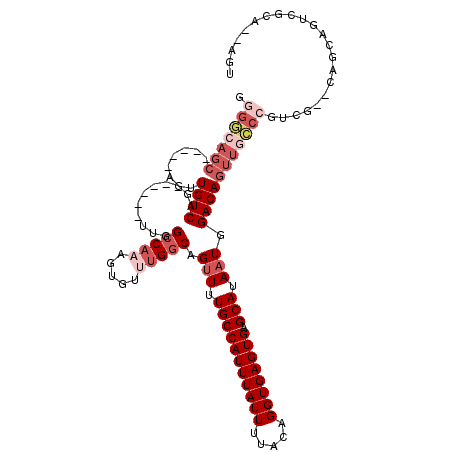
>2R_DroMel_CAF1 18516316 109 + 20766785 -GCAACCGGCAA-CCAGC-------AACUUGCAACUUGCGACUGCUGCGACUGUUG--CGACGGGCAACUGUCCAUUAUGCUCACUCACCUGUAAAAUAAAUGGCAAAGCUGCCAAACAC -(((....((((-(((((-------(..((((.....)))).))))).....))))--)(((((....))))).....)))....................(((((....)))))..... ( -32.20) >DroVir_CAF1 24181 91 + 1 -------AGCCACACAGC-------A---------UUGC---C--GUCAACAGCAGCCAGACGGCCAACUGUCCAUUAUGCUCACUCACCUGUAAAAUAAAUGGCAAAACUGCCA-ACAC -------.......(((.-------.---------((((---(--((....((((....(((((....))))).....)))).((......)).......)))))))..)))...-.... ( -18.90) >DroSec_CAF1 15281 116 + 1 -GCAACCGGCAA-CCAGCAACCAGCAACCAGCAACUUGCAACUGCUGUGACUGCUG--CGACGGGCAACUGUCCAUUAUGCUCACUCACCUGUAAAAUAAAUGGCAAAACUGCCAAACAC -......((((.-.(((((.((((((....((.....))...))))).)..)))))--.(((((....))))).....))))...................(((((....)))))..... ( -31.20) >DroEre_CAF1 18497 107 + 1 -GCAACCAGCAA-CCAGC-------AACCAGCAACUUGCAACU--UGCGACUGCUG--CGACGGGCAACUGUCCAUUAUGCUCACUCACCUGUAAAAUAAAUGGCAAAACUGCCAAACAC -(((..(((((.-...((-------((...((.....))...)--)))...)))))--.(((((....))))).....)))....................(((((....)))))..... ( -29.00) >DroYak_CAF1 21133 108 + 1 GGCAACUUGCAA-CCAGC-------AACUUGCAACUUGCAACU--CGCAACUGCUG--CGACGGGCAACUGUCCAUUAUGCUCACUCACCUGUAAAAUAAAUGGCAAAACUGCCAAACAC .((((.((((..-...))-------)).)))).....(((...--.(((.....))--)(((((....))))).....)))....................(((((....)))))..... ( -28.70) >consensus _GCAACCAGCAA_CCAGC_______AACCAGCAACUUGCAACU__UGCGACUGCUG__CGACGGGCAACUGUCCAUUAUGCUCACUCACCUGUAAAAUAAAUGGCAAAACUGCCAAACAC .......((((...................(((..(((((.....))))).))).....(((((....))))).....))))...................(((((....)))))..... (-16.98 = -17.82 + 0.84)

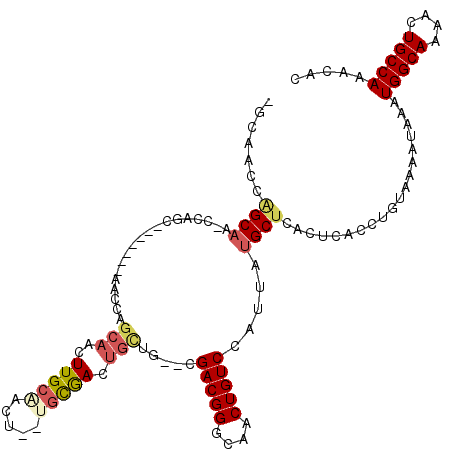
| Location | 18,516,347 – 18,516,451 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.65 |
| Mean single sequence MFE | -35.18 |
| Consensus MFE | -21.92 |
| Energy contribution | -24.03 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18516347 104 - 20766785 GGGGCAGC-----AGUUGUCAG------UUGGCCAAAGUGUUUGGCAGCUUUGCCAUUUAUUUUACAGGUGAGUGAGCAUAAUGGACAGUUGCCCGUCG--CAACAGUCGCAGCAGU .(((((((-----...((((((------(((.((((.....))))))))).(((((((((((.....)))))))).))).....)))))))))))((.(--(.......)).))... ( -35.80) >DroVir_CAF1 24197 109 - 1 GCGUGAGCCUCAGAGUUGUCAGCGC---GUUGCCAAAGUGU-UGGCAGUUUUGCCAUUUAUUUUACAGGUGAGUGAGCAUAAUGGACAGUUGGCCGUCUGGCUGCUGUUGAC--G-- ((....)).........(((((((.---((.((((..(.((-..((.(((((((((((((((.....)))))))).)))....)))).))..)))...)))).)))))))))--.-- ( -37.30) >DroSec_CAF1 15319 104 - 1 GGGGCAGC-----AGUUGUCAG------UUGGCCAAAGUGUUUGGCAGUUUUGCCAUUUAUUUUACAGGUGAGUGAGCAUAAUGGACAGUUGCCCGUCG--CAGCAGUCACAGCAGU .(((((((-----...((((.(------((((((((.....))))).....(((((((((((.....)))))))).))))))).)))))))))))((.(--(....)).))...... ( -33.70) >DroEre_CAF1 18528 102 - 1 GGGGCAGC-----AGUUGUCAG------UUGGCCAAAGUGUUUGGCAGUUUUGCCAUUUAUUUUACAGGUGAGUGAGCAUAAUGGACAGUUGCCCGUCG--CAGCAGUCGCA--AGU .(((((((-----...((((.(------((((((((.....))))).....(((((((((((.....)))))))).))))))).)))))))))))....--..((....)).--... ( -34.00) >DroWil_CAF1 22293 103 - 1 GGGA---------GUUUGUCAGCGCUGGCUGGGCCAAGUGU-UGGCAGUUUUGCCAUUUAUUUUACAGGUGAGUGAGCAUAAUAGACAGUUGCAAGUCU--CUUUUUUUGGG--GCC ....---------((((((((((((((((...))).)))))-)))).(((.(((((((((((.....)))))))).))).)))))))........((((--(.......)))--)). ( -32.30) >DroYak_CAF1 21165 102 - 1 GGGGCAGC-----AGUUGUCAG------UUGGCCAAAGUGUUUGGCAGUUUUGCCAUUUAUUUUACAGGUGAGUGAGCAUAAUGGACAGUUGCCCGUCG--CAGCAGUUGCG--AGU .(((((((-----...((((.(------((((((((.....))))).....(((((((((((.....)))))))).))))))).))))))))))).(((--(((...)))))--).. ( -38.00) >consensus GGGGCAGC_____AGUUGUCAG______UUGGCCAAAGUGUUUGGCAGUUUUGCCAUUUAUUUUACAGGUGAGUGAGCAUAAUGGACAGUUGCCCGUCG__CAGCAGUCGCA__AGU .(((((((........((((...........(((((.....))))).(((.(((((((((((.....)))))))).))).))).)))))))))))...................... (-21.92 = -24.03 + 2.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:32 2006