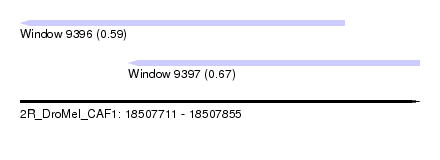
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,507,711 – 18,507,855 |
| Length | 144 |
| Max. P | 0.671949 |

| Location | 18,507,711 – 18,507,828 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.21 |
| Mean single sequence MFE | -28.85 |
| Consensus MFE | -14.64 |
| Energy contribution | -15.52 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.590981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18507711 117 - 20766785 CUAUCAACAAGAAUUUAGACUCUAUCUUAUCUGCAC--AUGAGUGGUUUAGAUGAGUCCCAGUUCUGAAGUGGCCAAUUCAAUGAAUUACGACCUUAUCUGAUC-UACAGAUAGGCAGAC ....((...((((((..(((((.((((...(..(..--....)..)...)))))))))..))))))....))((((((((...))))).......((((((...-..))))))))).... ( -30.10) >DroSec_CAF1 6711 117 - 1 CUAUCAACGUGAAUUUAGACUCUAUCUAAUCUGCAC--AUGAGUGGGUUAGAUGAGUUCCAGUUCUGAAGUGGCCAAUUCAAUGAAUUACGACGGUAUCUGAUC-UACAGAUAGGCAGAC ....((....(((((..(((((.((((((((..(..--....)..)))))))))))))..))))).....))((((((((...))))).......((((((...-..))))))))).... ( -33.00) >DroSim_CAF1 4902 117 - 1 CUAUCAACGUGAAUUUAGACUCUAUCUAAUCUGCAC--AUGAGUGGGUUAGAUGAGUUCCAGUUCUGAAGUGGCCAAUUCAAUGAAUUACGACCUUAUCUGAUC-UACAGAUAGGCAGAC (((((...(((...(((((.....)))))....)))--....((((((((((((((.((.(((((((((........))))..)))))..)).)))))))))))-))).)))))...... ( -35.00) >DroEre_CAF1 9714 106 - 1 CCAUCAUCGUGAAUUUAUACACUAUCUAAUCUG-------------GUUAGAUGAAUUCCAGUUCUAAAGUGGCCAACUCAAUGAAUUACGACCUUAUCUGAUC-UACAGAUGGUCAGAC (((((...(((........)))..........(-------------(((((((((..((.(((((...(((.....)))....)))))..))..))))))))))-....)))))...... ( -22.20) >DroYak_CAF1 12115 117 - 1 CCAUCGACGUGAGUUUAAACACUAUCUAAUCUGCAA--CCGAGUGGGUUAGAUGAGUCCCAGCUCUGAAGUGGCCAAUUCAUUGAAUUACGACCUUAUCUGAUC-UACAGAUAGUCAGAC .....((((((((((....((((((((((((..(..--....)..))))))))((((....))))...))))...))))))).............((((((...-..))))))))).... ( -32.00) >DroPer_CAF1 4636 98 - 1 ------------------GGAGUAUCUAAUCUAGACAGAGAAAUGGGUUUGAUAAGUCUCGCUCUUCGAGUGGCCUCUUGAAUGAAUUACGG----AUCUACUCGUAUAGAUAGACAGUC ------------------....((((.((((((..........)))))).)))).((((..((...(((((((..((.(((.....))).))----..)))))))...))..)))).... ( -20.80) >consensus CUAUCAACGUGAAUUUAGACACUAUCUAAUCUGCAC__AUGAGUGGGUUAGAUGAGUCCCAGUUCUGAAGUGGCCAAUUCAAUGAAUUACGACCUUAUCUGAUC_UACAGAUAGGCAGAC ......................((((((((((((........)))))))))))).(((..(((((((((........))))..)))))..)))..((((((......))))))....... (-14.64 = -15.52 + 0.88)

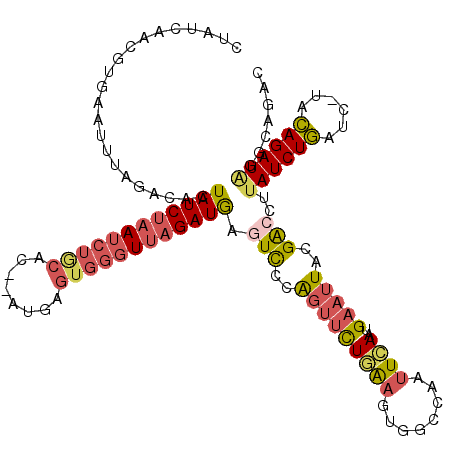
| Location | 18,507,750 – 18,507,855 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.84 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -17.89 |
| Energy contribution | -19.74 |
| Covariance contribution | 1.85 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18507750 105 - 20766785 GCCAGACACA-------------ACUUCCAAUAAAAUCGACUAUCAACAAGAAUUUAGACUCUAUCUUAUCUGCACAUGAGUGGUUUAGAUGAGUCCCAGUUCUGAAGUGGCCAAUUC (((.......-------------(((((..........(.....)....((((((..(((((.((((...(..(......)..)...)))))))))..))))))))))))))...... ( -22.91) >DroSec_CAF1 6750 105 - 1 GCCAGACACA-------------ACAUCCCAUUAAAUCGACUAUCAACGUGAAUUUAGACUCUAUCUAAUCUGCACAUGAGUGGGUUAGAUGAGUUCCAGUUCUGAAGUGGCCAAUUC ((((......-------------...............(.....)..((.(((((..(((((.((((((((..(......)..)))))))))))))..)))))))...))))...... ( -25.80) >DroSim_CAF1 4941 105 - 1 GCCAGACACA-------------ACUUCCAAUUAAAUCAACUAUCAACGUGAAUUUAGACUCUAUCUAAUCUGCACAUGAGUGGGUUAGAUGAGUUCCAGUUCUGAAGUGGCCAAUUC (((.......-------------(((((...........((.......))(((((..(((((.((((((((..(......)..)))))))))))))..))))).))))))))...... ( -26.11) >DroEre_CAF1 9753 107 - 1 GCCAGACACAGUUUACACGGCACACUUCCAAUUAAGUGGACCAUCAUCGUGAAUUUAUACACUAUCUAAUCUG-----------GUUAGAUGAAUUCCAGUUCUAAAGUGGCCAACUC ..................(((.(((((......((.((((...((((((((........))).(((......)-----------))..)))))..)))).))...))))))))..... ( -21.50) >DroYak_CAF1 12154 118 - 1 GCCAGACACAAUUUACAAGGCACACUUGCAAUUAAAUCGACCAUCGACGUGAGUUUAAACACUAUCUAAUCUGCAACCGAGUGGGUUAGAUGAGUCCCAGCUCUGAAGUGGCCAAUUC ..................(((.(((((.((......(((.....)))...(((((.....(((((((((((..(......)..)))))))).)))...)))))))))))))))..... ( -30.60) >consensus GCCAGACACA_____________ACUUCCAAUUAAAUCGACUAUCAACGUGAAUUUAGACUCUAUCUAAUCUGCACAUGAGUGGGUUAGAUGAGUUCCAGUUCUGAAGUGGCCAAUUC ((((..................................(.....).....(((((..(((((.(((((((((((......))))))))))))))))..))))).....))))...... (-17.89 = -19.74 + 1.85)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:26 2006