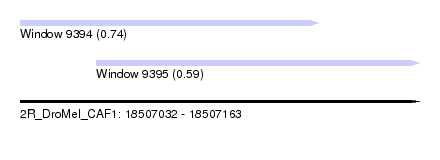
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,507,032 – 18,507,163 |
| Length | 131 |
| Max. P | 0.743705 |

| Location | 18,507,032 – 18,507,130 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 85.85 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -21.14 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18507032 98 + 20766785 --ACCGACG-CCCAGAUUUGUUUGCUAUAUGCCAAUUUGGCGGUGUGUUUGCAUAUUUACUGUAAACACUGUCCAAAUUAGGCGGAGAUUUCCAUUCACCG- --..(..((-((..((((((.........((((.....))))..(((((((((.......)))))))))....)))))).))))..)..............- ( -26.30) >DroPse_CAF1 3902 101 + 1 UUGCCUGAGUUUCUGUUCUGUUUGUUCUAUGCCAAUUUAGCGGCUUGUUUGCAUAUUUACUGUACACACUGUCUAAAUUAGGCGGAGAUUUCCUUUCAGCC- ....(((((.....................(((((((((((((..(((.((((.......)))).)))))).))))))).)))((......)).)))))..- ( -21.40) >DroSim_CAF1 3517 98 + 1 --AACUCCG-CCCAGAUUUGUUUGCUAUAUGCCAAUUUGGCGGUCUGUUUGCAUAUUUACUGUAAACACUGUCCAAAUUAGGCGGAGAUUUCCAUUCACCG- --..(((((-((...........((.....)).(((((((((((..(((((((.......))))))))))).))))))).)))))))..............- ( -31.20) >DroEre_CAF1 9094 100 + 1 CCACCUCCG-CCCAGAUUUGUUUGCUAUAUGCCAAUUUGGCGGUCUGUUUGCAUAUUUACUGUAAACACUGUCCAAAUUAGGCGAAGAUUUCCAUUCAGCG- .........-.............(((....((((((((((((((..(((((((.......))))))))))).))))))).)))(((........)))))).- ( -25.80) >DroYak_CAF1 11482 101 + 1 CCACCUCAG-CCCAGAUUUGUUUGCUAUAUGCCAAUUUGGCGGUCUGUUUGCAUAUUUACUGUAAACACUGUCCAAAUUAGGCGGCGCUUUCCAUUCACCGC ........(-((((((....))))......((((((((((((((..(((((((.......))))))))))).))))))).))))))................ ( -26.70) >DroPer_CAF1 3812 101 + 1 UUGCCUGAGUUUCUGUUCUGUUUGUUCUAUGCCAAUUUAGCGGCUUGUUUGCAUAUUUACUGUACACACUGUCUAAAUUAGGCGGAGAUUUCCUUUCAGCC- ....(((((.....................(((((((((((((..(((.((((.......)))).)))))).))))))).)))((......)).)))))..- ( -21.40) >consensus __ACCUCAG_CCCAGAUUUGUUUGCUAUAUGCCAAUUUGGCGGUCUGUUUGCAUAUUUACUGUAAACACUGUCCAAAUUAGGCGGAGAUUUCCAUUCACCG_ ..............................(((((((((((((..((((((((.......))))))))))).))))))).)))((......))......... (-21.14 = -21.20 + 0.06)


| Location | 18,507,057 – 18,507,163 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 85.08 |
| Mean single sequence MFE | -34.73 |
| Consensus MFE | -23.92 |
| Energy contribution | -24.42 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18507057 106 + 20766785 AUGCCAAUUUGGCGGUGUGUUUGCAUAUUUACUGUAAACACUGUCCAAAUUAGGCGGAGAUUUCCAUUCACCG---CCGUCCUUGA---GCU--AUUUGCCUAGGGGGCAUUCC ((((((((((((((((((..(((((.......))))))))))).))))))).(((((.((.......)).)))---))..(((((.---((.--....)).))))))))))... ( -41.20) >DroPse_CAF1 3930 109 + 1 AUGCCAAUUUAGCGGCUUGUUUGCAUAUUUACUGUACACACUGUCUAAAUUAGGCGGAGAUUUCCUUUCAGCC---AUGUCCUUGGCACCUU--GUUUGUCCCUGGGAAAUACC ..(((((((((((((..(((.((((.......)))).)))))).))))))).)))((..(((((((....(((---(......))))..(..--....).....))))))).)) ( -27.30) >DroSec_CAF1 6080 106 + 1 AUGCCAAUUUGGCGGUCUGUUUGCAUAUUUACUGUAAACACUGUCCAAAUUAGGCGGAGGUUUCCAUUCACCG---CCGUCCUUGA---GUU--GUUUGCGCAAGGGGCAUUCC ((((((((((((((((..(((((((.......))))))))))).))))))).(((((.((.......)).)))---))..(((((.---((.--....)).))))))))))... ( -40.10) >DroEre_CAF1 9121 106 + 1 AUGCCAAUUUGGCGGUCUGUUUGCAUAUUUACUGUAAACACUGUCCAAAUUAGGCGAAGAUUUCCAUUCAGCG---CCGUCCUUGA---GUU--AUUUGCCCAAGGGGAAUUCC ...(((((((((((((..(((((((.......))))))))))).))))))).((((..((.......))..))---))..(((((.---((.--....)).)))))))...... ( -32.40) >DroYak_CAF1 11509 111 + 1 AUGCCAAUUUGGCGGUCUGUUUGCAUAUUUACUGUAAACACUGUCCAAAUUAGGCGGCGCUUUCCAUUCACCGCCGCCGUCCUUGA---GCUGAGUUUGCCCAAGGGGAAUUCC ...(((((((((((((..(((((((.......))))))))))).))))))).(((((((............)))))))..(((((.---((.......)).)))))))...... ( -40.10) >DroPer_CAF1 3840 109 + 1 AUGCCAAUUUAGCGGCUUGUUUGCAUAUUUACUGUACACACUGUCUAAAUUAGGCGGAGAUUUCCUUUCAGCC---AUGUCCUUGGCACCUU--GUUUGUCCCUGGGAAAUACC ..(((((((((((((..(((.((((.......)))).)))))).))))))).)))((..(((((((....(((---(......))))..(..--....).....))))))).)) ( -27.30) >consensus AUGCCAAUUUGGCGGUCUGUUUGCAUAUUUACUGUAAACACUGUCCAAAUUAGGCGGAGAUUUCCAUUCACCG___CCGUCCUUGA___GUU__GUUUGCCCAAGGGGAAUUCC ..(((((((((((((..((((((((.......))))))))))).))))))).)))((......))..............((((((................))))))....... (-23.92 = -24.42 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:24 2006