
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,988,911 – 2,989,012 |
| Length | 101 |
| Max. P | 0.664900 |

| Location | 2,988,911 – 2,989,012 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.50 |
| Mean single sequence MFE | -31.87 |
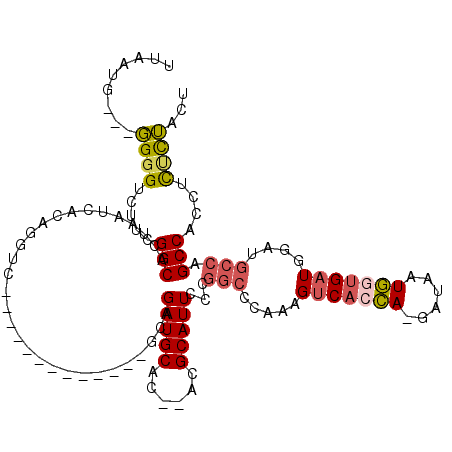
| Consensus MFE | -19.84 |
| Energy contribution | -20.15 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
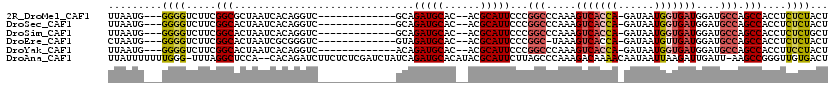
>2R_DroMel_CAF1 2988911 101 + 20766785 UUAAUG---GGGGUCUUCGGCGCUAAUCACAGGUC-------------GCAGAUGCAC--ACGCAUUCCCGGCCCAAAGUCACCA-GAUAAUGGUGAUGGAUGCCAGCCACCUCUCUACU .....(---(((((....(((....(((...((((-------------(..(((((..--..)))))..)))))....(((((((-.....))))))).)))....)))))))))..... ( -36.00) >DroSec_CAF1 13488 101 + 1 UUAAUG---GGGGUCUUCGGCACUAAUCACAGGUC-------------GCAGAUGCAC--ACGCAUUCCCGGCCCAAAGUCACCA-GAUAAUGGUGAUGGAUGCCAGCCACCUCUCUACU .....(---(((((....((((((.......((((-------------(..(((((..--..)))))..)))))....(((((((-.....))))))))).))))....))))))..... ( -35.70) >DroSim_CAF1 13487 101 + 1 UUAAUG---GGGGUCUUCGGCACUAAUCACAGGUC-------------GCAGAUGCAC--ACGCAUUCCCGGCCCAAAGUCACCA-GAUAAUGGUGAUGGAUGCCAGCCACCUCUCUGCU .....(---(((((....((((((.......((((-------------(..(((((..--..)))))..)))))....(((((((-.....))))))))).))))....))))))..... ( -35.70) >DroEre_CAF1 13873 100 + 1 CUAAUG---GGGGUCUUCGGCACUAAUCGCGGGUC-------------GUAGAUGCAC--ACGCAUUCCCGGC-UAAAGUCACCA-GAUAAUGUUGAUGGAUGCCAGCCACCUCUCUACU .....(---(((((....(((.........(((..-------------...(((((..--..))))))))(((-....((((.((-.....)).))))....))).)))))))))..... ( -28.70) >DroYak_CAF1 13901 101 + 1 UUAAUG---GGGGUCUUCGGCACUAAUCACAGGUC-------------ACAGAUGCAC--ACGCAUUCCCGGCCCAAAGUCACCA-GAUAAUGGUGAUGGAUGCCAGCCACCUUCCUACU .....(---(((((....((((((.......((((-------------...(((((..--..)))))...))))....(((((((-.....))))))))).))))....))))))..... ( -31.40) >DroAna_CAF1 12777 116 + 1 UUAUUUUUUUGGG-UUUAGGCUCCA--CACAGAUCUUCUCUCGAUCUAUCAGAUGCACAUACGCAUUCUUAGCCCAAAGACAAAACAAUAAUUAAGAUUGAUU-AAGCCGGGUUGUGACU .....((((((((-((.((......--...(((((.......)))))....(((((......))))))).))))))))))....(((((..((((......))-)).....))))).... ( -23.70) >consensus UUAAUG___GGGGUCUUCGGCACUAAUCACAGGUC_____________GCAGAUGCAC__ACGCAUUCCCGGCCCAAAGUCACCA_GAUAAUGGUGAUGGAUGCCAGCCACCUCUCUACU .........((((.....(((..............................(((((......)))))...(((.....(((((((......)))))))....))).)))....))))... (-19.84 = -20.15 + 0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:10 2006