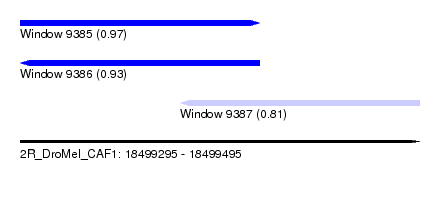
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,499,295 – 18,499,495 |
| Length | 200 |
| Max. P | 0.968835 |

| Location | 18,499,295 – 18,499,415 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -32.83 |
| Energy contribution | -33.83 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
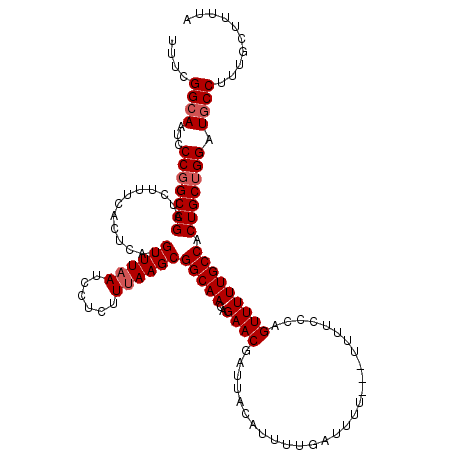
>2R_DroMel_CAF1 18499295 120 + 20766785 GCACCCAGUUGUUGUAUGGUAUGCGAUAGGAUUUUUAUGUAAAACAUAAUUGCAGAUUAGUCGCGGCGGCAAUGUGCAAAUAAAAGCAAAGGCAUCCAGCAGUGGCAAAAACUGAGAAAA .......((..((((.(((.((((..........(((((.....)))))(((((.(((.((((...))))))).)))))............)))))))))))..)).............. ( -29.10) >DroSec_CAF1 18098 120 + 1 GCACCCAGUUGUUGUAUGGGAUGCGAUAGGAUUUUUAUGUAAAACAUAAUUGCAGAUUAGUCGCGGCGGCAUUGUGCAAAUAAAAGCAAAGGCAUCCAGCAGUGGCAAAAACUGGGAAAA ...(((((((.((((.((((((((..........(((((.....)))))(((((.(...((((...))))..).)))))............))))))..))...)))).))))))).... ( -36.00) >DroSim_CAF1 7136 120 + 1 GCACCCAGUUGUUGUAUGGGAUGCGAUAGGAUUUUUAUGUAAAACAUAAUUGCAGAUUAGUCGCGGCGGCAAUGUGCAAAUAAAAGCAAAGGCAUCCUGCAGUGGCAAAAACUGGGAAAA ...(((((((.((((.((((((((..........(((((.....)))))(((((.(((.((((...))))))).)))))............)))))))).....)))).))))))).... ( -38.70) >consensus GCACCCAGUUGUUGUAUGGGAUGCGAUAGGAUUUUUAUGUAAAACAUAAUUGCAGAUUAGUCGCGGCGGCAAUGUGCAAAUAAAAGCAAAGGCAUCCAGCAGUGGCAAAAACUGGGAAAA ...(((((((.((((.((((((((..........(((((.....)))))(((((.(((.((((...))))))).)))))............))))))..))...)))).))))))).... (-32.83 = -33.83 + 1.00)

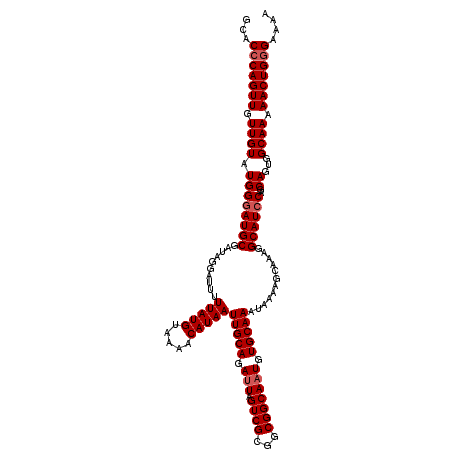

| Location | 18,499,295 – 18,499,415 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -26.19 |
| Energy contribution | -26.30 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18499295 120 - 20766785 UUUUCUCAGUUUUUGCCACUGCUGGAUGCCUUUGCUUUUAUUUGCACAUUGCCGCCGCGACUAAUCUGCAAUUAUGUUUUACAUAAAAAUCCUAUCGCAUACCAUACAACAACUGGGUGC ....(((((((.(((....((.(((((((..((((..(((.((((...........)))).)))...))))(((((.....)))))..........)))).)))))))).)))))))... ( -23.40) >DroSec_CAF1 18098 120 - 1 UUUUCCCAGUUUUUGCCACUGCUGGAUGCCUUUGCUUUUAUUUGCACAAUGCCGCCGCGACUAAUCUGCAAUUAUGUUUUACAUAAAAAUCCUAUCGCAUCCCAUACAACAACUGGGUGC ....(((((((.(((....((..((((((..((((..(((.((((...........)))).)))...))))(((((.....)))))..........))))))))..))).)))))))... ( -29.20) >DroSim_CAF1 7136 120 - 1 UUUUCCCAGUUUUUGCCACUGCAGGAUGCCUUUGCUUUUAUUUGCACAUUGCCGCCGCGACUAAUCUGCAAUUAUGUUUUACAUAAAAAUCCUAUCGCAUCCCAUACAACAACUGGGUGC ....(((((((..(((....)))((((((..((((..(((.((((...........)))).)))...))))(((((.....)))))..........))))))........)))))))... ( -29.80) >consensus UUUUCCCAGUUUUUGCCACUGCUGGAUGCCUUUGCUUUUAUUUGCACAUUGCCGCCGCGACUAAUCUGCAAUUAUGUUUUACAUAAAAAUCCUAUCGCAUCCCAUACAACAACUGGGUGC ....(((((((.(((....((..((((((..((((..(((.((((...........)))).)))...))))(((((.....)))))..........))))))))..))).)))))))... (-26.19 = -26.30 + 0.11)

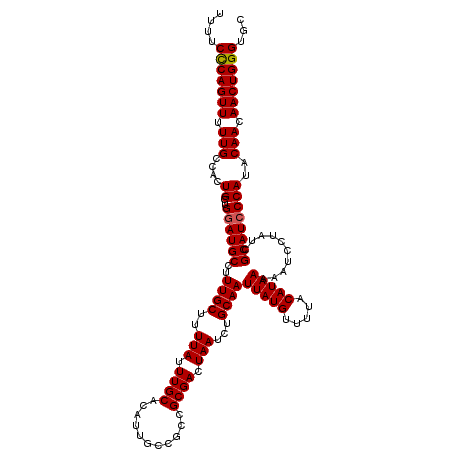
| Location | 18,499,375 – 18,499,495 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.08 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -23.36 |
| Energy contribution | -24.36 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18499375 120 - 20766785 UUUCGGGAAUCCCGGCAGCUCUUUCACUCAUGUUUAAUCCUCUUCAAGCGGCAACAGAACGAUUACAUUUUGAUUUUUUUUUUUCUCAGUUUUUGCCACUGCUGGAUGCCUUUGCUUUUA ....(((.((((.(((((.............((((.(......).))))(((((..((((............................))))))))).))))))))).)))......... ( -21.19) >DroSec_CAF1 18178 117 - 1 UUUCGGCAAUCCCGGCAGCUCUUUCACUCAUGUUUAAUCCUCUUUAAGCGGCAACAGAACGAUUACAUUUUGAUUUU---UUUUCCCAGUUUUUGCCACUGCUGGAUGCCUUUGCUUUUA ....((((...(((((((.............((((((......))))))(((((..((((.................---........))))))))).))))))).)))).......... ( -28.21) >DroSim_CAF1 7216 117 - 1 UUUCGGCAAUCCCGGCAGCUCUUUCACUCAUGUUUAAUCCUCUUUAAGCGGCAACAGAACGAUUACAUUUUGAUUUU---UUUUCCCAGUUUUUGCCACUGCAGGAUGCCUUUGCUUUUA ....((((.(((..((((.............((((((......))))))(((((..((((.................---........))))))))).)))).))))))).......... ( -26.21) >consensus UUUCGGCAAUCCCGGCAGCUCUUUCACUCAUGUUUAAUCCUCUUUAAGCGGCAACAGAACGAUUACAUUUUGAUUUU___UUUUCCCAGUUUUUGCCACUGCUGGAUGCCUUUGCUUUUA ....((((...(((((((.............((((((......))))))(((((..((((............................))))))))).))))))).)))).......... (-23.36 = -24.36 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:16 2006