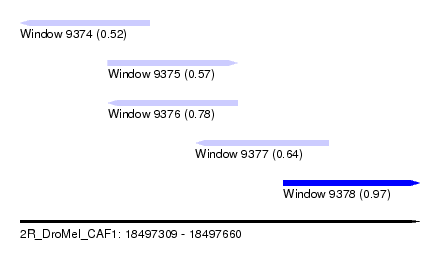
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,497,309 – 18,497,660 |
| Length | 351 |
| Max. P | 0.972255 |

| Location | 18,497,309 – 18,497,423 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.01 |
| Mean single sequence MFE | -29.34 |
| Consensus MFE | -24.50 |
| Energy contribution | -24.81 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18497309 114 - 20766785 GCUAAUUCAGACCUAGGCCAGCUAGGUUCAGCAGCCU---CCCAGCCU-UUCAG--CUGCUCUCCCCUCGCAUUACGUAUACGCCCCUUAUGCCACAUAUCACGUAUACGCAGUGCUUGA .........(((((((.....))))))).((((((..---........-....)--)))))........(((((.((((((((.....((((...))))...)))))))).))))).... ( -31.44) >DroSec_CAF1 15938 113 - 1 GCUAAUUCAGACCUAGGCCGUCUAGGUUCAGCAGCCU---CCCAGCCU-UUCAG--CGGCUCUCCCCUCGCAUUACGUAUACGCCCCU-CUGCCACAUAUCACGUAUACGCAGUGCUCGA (((......(((((((.....)))))))....)))..---...((((.-.....--.))))........(((((.((((((((.....-.............)))))))).))))).... ( -28.27) >DroSim_CAF1 5089 114 - 1 GCUAAUUCAGACCUAGGCCAGCUAGGUUCAGCAGCCUU--CCCAGCCA-UUCAG--CGGCUCUCCCCUCGCAUUACGUAUACGCCCCU-CUGCCACAUAUCACGUAUACGCAGUGCUCGA (((......(((((((.....)))))))....)))...--...((((.-.....--.))))........(((((.((((((((.....-.............)))))))).))))).... ( -28.57) >DroEre_CAF1 21505 119 - 1 GCUAAUUCAGGCCUAGACCAGCUAGGUUCAGCAGCCUUGCCUCAGCCGCUUCCGCCCAGCACUCCCCUCGCAUUACGUAUACGCCCCG-CUGCCACAUAUCACGUAUACGCAGUGCUUGA (((......(((..((....((.((((......)))).))))..)))((....))..))).........(((((.((((((((.....-.............)))))))).))))).... ( -29.07) >consensus GCUAAUUCAGACCUAGGCCAGCUAGGUUCAGCAGCCU___CCCAGCCU_UUCAG__CGGCUCUCCCCUCGCAUUACGUAUACGCCCCU_CUGCCACAUAUCACGUAUACGCAGUGCUCGA (((......(((((((.....)))))))....))).........(((..........))).........(((((.((((((((...................)))))))).))))).... (-24.50 = -24.81 + 0.31)

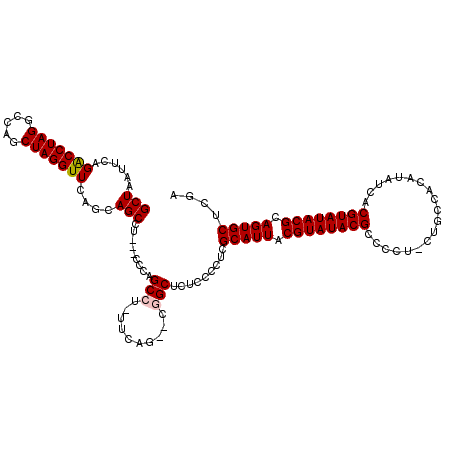
| Location | 18,497,386 – 18,497,500 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.78 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -22.33 |
| Energy contribution | -22.57 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18497386 114 + 20766785 ---AGGCUGCUGAACCUAGCUGGCCUAGGUCUGAAUUAGCAUAAUAUGCCAAAAUGGCAGUUUUUUAAGAAUUCGCCAUAAGUGU---UGGGUUUCUCAACCAAAAAGUGCCAGCCUUUG ---(((((((((....)))).(((((.(((..(((((.........((((.....))))..........))))))))....(.((---((((...)))))))....)).))))))))... ( -31.21) >DroSec_CAF1 16014 115 + 1 ---AGGCUGCUGAACCUAGACGGCCUAGGUCUGAAUUAGCAUAAUAUGCCAUAAUGGCAGUUUUUUAACAAUUCGCCAUAAGUGUUGUUGGGUUUCUAAGCCAAAA--GGACUGCCUUUG ---.(((((((((((((((.....)))))).....))))))......))).....(((((((((((..((((.(((.....)))..))))((((....)))).)))--)))))))).... ( -36.20) >DroSim_CAF1 5165 116 + 1 --AAGGCUGCUGAACCUAGCUGGCCUAGGUCUGAAUUAGCAUAAUAUGCCAUAAUGGCAGUUUUUUAACAAUUCGCCAUAAGUGUUGUUGGGUUUCUAAGCCAAAG--GGCCUGCCUUUG --..((((...(((((((((.(((((.(((..(((((.........((((.....))))(((....)))))))))))...)).))))))))))))...)))).(((--((....))))). ( -34.80) >DroEre_CAF1 21584 114 + 1 GCAAGGCUGCUGAACCUAGCUGGUCUAGGCCUGAAUUAGCAUAAUAUGCCAUAAUGGCAGUUUUUUAACAAUAC-CCAUAAGUGU---UGGAUUUGAAAACCGAAG--GGUCUGCCUUUG ((.(((((((((....)))).)))))((((((..............((((.....))))((((((...((((((-......))))---)).....))))))....)--)))))))..... ( -30.60) >consensus ___AGGCUGCUGAACCUAGCUGGCCUAGGUCUGAAUUAGCAUAAUAUGCCAUAAUGGCAGUUUUUUAACAAUUCGCCAUAAGUGU___UGGGUUUCUAAACCAAAA__GGCCUGCCUUUG ...((((((((((((((((.....)))))).....)))))).....((((.....))))...............................((((....))))...........))))... (-22.33 = -22.57 + 0.25)

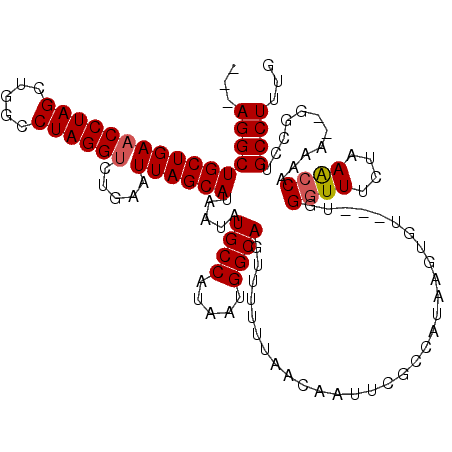
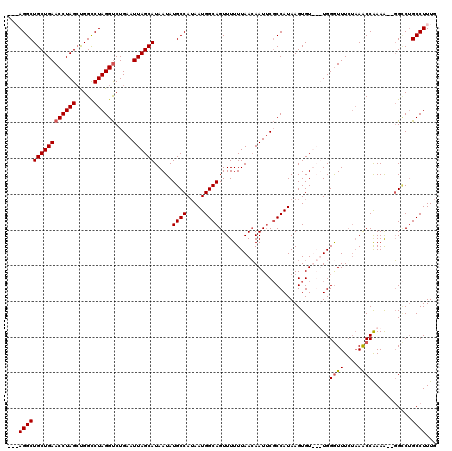
| Location | 18,497,386 – 18,497,500 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.78 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -22.16 |
| Energy contribution | -22.97 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18497386 114 - 20766785 CAAAGGCUGGCACUUUUUGGUUGAGAAACCCA---ACACUUAUGGCGAAUUCUUAAAAAACUGCCAUUUUGGCAUAUUAUGCUAAUUCAGACCUAGGCCAGCUAGGUUCAGCAGCCU--- ....(((((((.((....((((..(((.....---......((((((..............)))))).(((((((...)))))))))).)))).)))))))))((((......))))--- ( -32.44) >DroSec_CAF1 16014 115 - 1 CAAAGGCAGUCC--UUUUGGCUUAGAAACCCAACAACACUUAUGGCGAAUUGUUAAAAAACUGCCAUUAUGGCAUAUUAUGCUAAUUCAGACCUAGGCCGUCUAGGUUCAGCAGCCU--- ...((((.((((--(..((((((((................((((((..((......))..))))))..((((((...))))))........))))))))...)))....)).))))--- ( -29.50) >DroSim_CAF1 5165 116 - 1 CAAAGGCAGGCC--CUUUGGCUUAGAAACCCAACAACACUUAUGGCGAAUUGUUAAAAAACUGCCAUUAUGGCAUAUUAUGCUAAUUCAGACCUAGGCCAGCUAGGUUCAGCAGCCUU-- ..(((((.(..(--(((((((((((................((((((..((......))..))))))..((((((...))))))........)))))))))..)))..)....)))))-- ( -31.10) >DroEre_CAF1 21584 114 - 1 CAAAGGCAGACC--CUUCGGUUUUCAAAUCCA---ACACUUAUGG-GUAUUGUUAAAAAACUGCCAUUAUGGCAUAUUAUGCUAAUUCAGGCCUAGACCAGCUAGGUUCAGCAGCCUUGC ....((((((((--....)))...((((((((---.......)))-)).)))........)))))....((((((...))))))....((((((..(((.....)))..))..))))... ( -27.60) >consensus CAAAGGCAGGCC__CUUUGGCUUAGAAACCCA___ACACUUAUGGCGAAUUGUUAAAAAACUGCCAUUAUGGCAUAUUAUGCUAAUUCAGACCUAGGCCAGCUAGGUUCAGCAGCCU___ ...((((...........(((((((................((((((..((......))..))))))..((((((...))))))........))))))).(((......))).))))... (-22.16 = -22.97 + 0.81)

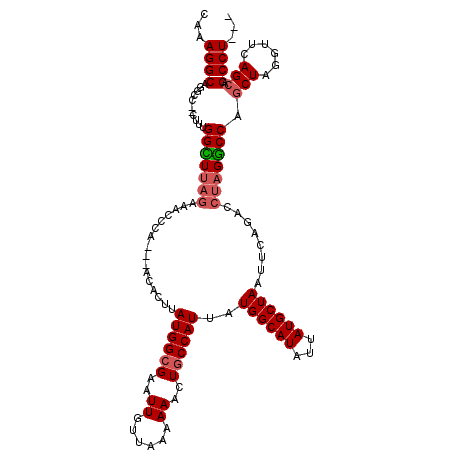

| Location | 18,497,463 – 18,497,580 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.31 |
| Mean single sequence MFE | -21.18 |
| Consensus MFE | -15.99 |
| Energy contribution | -16.05 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18497463 117 - 20766785 GAACAGAUCCAUUCAGUCUGGCCAAAAAGUUAUUAAAUUAUUUUAAAAUCAAACCAUCCAUCCAUUGCACUGCGCAGCCACAAAGGCUGGCACUUUUUGGUUGAGAAACCCA---ACACU ...(((((.......)))))((((((((((..(((((....))))).........................((.(((((.....)))))))))))))))))...........---..... ( -26.70) >DroSec_CAF1 16091 118 - 1 GAACAGACCCAUUCUGUCUGGCCAAAAAGUUAUUAAAUUAUUUUAAGAUCAAACCAUCCAUCCAUUGCACUGCGCAGCCACAAAGGCAGUCC--UUUUGGCUUAGAAACCCAACAACACU ...(((((.......)))))(((((((.....(((((....)))))(((................(((.....)))(((.....))).))).--)))))))................... ( -20.60) >DroSim_CAF1 5243 118 - 1 GAACAGACCCAUUCUGUCUGGCCAAAAAGUUAUUAAAUUAUUUUAAAAUCAAACCAUCCAUCCAUUGCACUGCGCAGCCACAAAGGCAGGCC--CUUUGGCUUAGAAACCCAACAACACU ...(((((.......)))))((((((......(((((....))))).......((..........(((.....)))(((.....))).))..--.))))))................... ( -21.10) >DroEre_CAF1 21663 112 - 1 GAACAGACCCAUUUUGUCUGGCCAAAAAGUUAUUAAAUUAUUUUAAAAUCAAACCAUCCAU---UUGCACUGCACAGCCACAAAGGCAGACC--CUUCGGUUUUCAAAUCCA---ACACU (..(((((.......)))))..).....(((.(((((....)))))....(((((......---.(((...)))..(((.....))).....--....)))))........)---))... ( -16.30) >consensus GAACAGACCCAUUCUGUCUGGCCAAAAAGUUAUUAAAUUAUUUUAAAAUCAAACCAUCCAUCCAUUGCACUGCGCAGCCACAAAGGCAGGCC__CUUUGGCUUAGAAACCCA___ACACU ...(((((.......)))))((((((.......................................(((.....)))(((.....)))........))))))................... (-15.99 = -16.05 + 0.06)

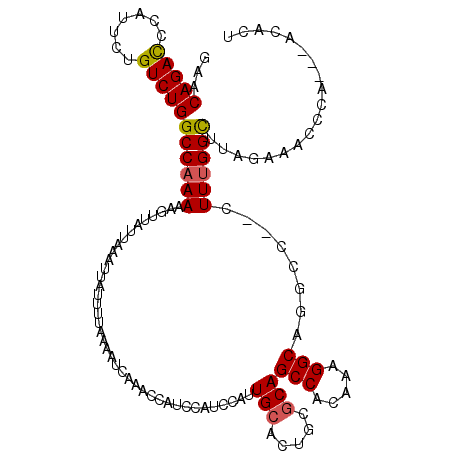
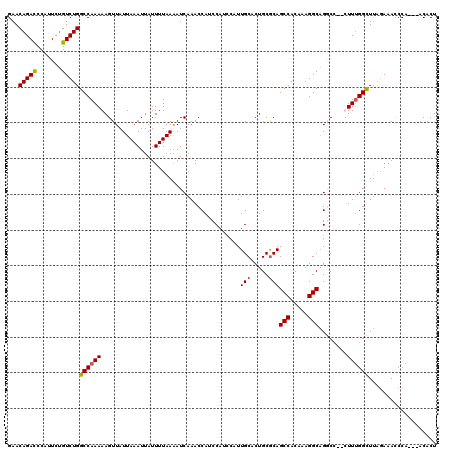
| Location | 18,497,540 – 18,497,660 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -35.42 |
| Energy contribution | -35.17 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18497540 120 + 20766785 UAAUUUAAUAACUUUUUGGCCAGACUGAAUGGAUCUGUUCAAUGCAGGCGGGCCGAAGAAGGGAUUGACAAGGCGGAAAACAUUUUGCCUGGCAAGGCCUAAAACACGCUUGAAUUAUUC ((((((((....(((((((((...(((..((((....))))...)))...)))))))))(((..(((...((((((((....))))))))..)))..))).........))))))))... ( -36.60) >DroSec_CAF1 16169 120 + 1 UAAUUUAAUAACUUUUUGGCCAGACAGAAUGGGUCUGUUCAAGGCAGGCGGGCCGAAGAAGGGAUUGACAAGGCGGAAAACAUUUUGCCUGGCAAGGCCUAAAACACGCUUGAAUUAUUC ((((((((....(((((((((...(.....).((((((.....)))))).)))))))))(((..(((...((((((((....))))))))..)))..))).........))))))))... ( -36.70) >DroSim_CAF1 5321 120 + 1 UAAUUUAAUAACUUUUUGGCCAGACAGAAUGGGUCUGUUCAAGGCAGGCGGGCCAAAGAAGGGAUUGACAAGGCGGAAAACAUUUUGCCUGGCAAGGCCUAAAACACGCUUGAAUUAUUC ((((((((....(((((((((...(.....).((((((.....)))))).)))))))))(((..(((...((((((((....))))))))..)))..))).........))))))))... ( -37.00) >DroEre_CAF1 21735 120 + 1 UAAUUUAAUAACUUUUUGGCCAGACAAAAUGGGUCUGUUCAAUGCAGGCGGGCCGAGAAGGGGAUUGACAAGGCGGAAAACAUUUUGCCCGGCAAGGCCUAAAACACGCUUGAAUUAUUC ((((((((...((((((((((...(.....).((((((.....)))))).))))))))))(((.(((.(..(((((((....)))))))..))))..))).........))))))))... ( -37.90) >consensus UAAUUUAAUAACUUUUUGGCCAGACAGAAUGGGUCUGUUCAAGGCAGGCGGGCCGAAGAAGGGAUUGACAAGGCGGAAAACAUUUUGCCUGGCAAGGCCUAAAACACGCUUGAAUUAUUC ((((((((....(((((((((...(.....).((((((.....)))))).)))))))))(((..(((...((((((((....))))))))..)))..))).........))))))))... (-35.42 = -35.17 + -0.25)

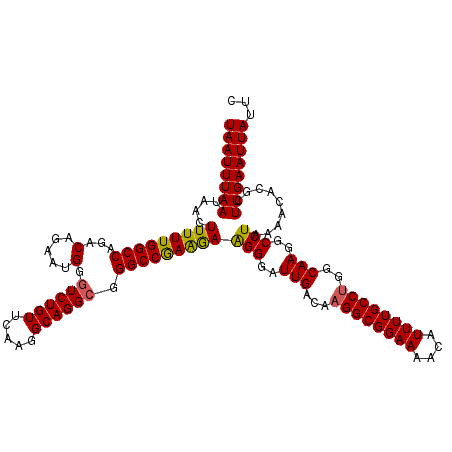
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:56:08 2006