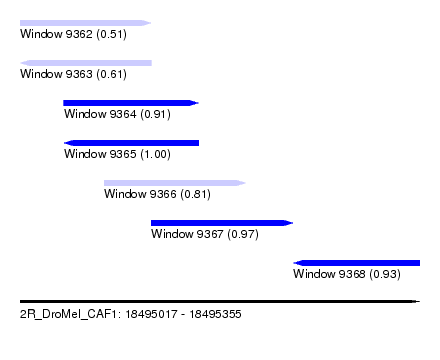
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,495,017 – 18,495,355 |
| Length | 338 |
| Max. P | 0.999928 |

| Location | 18,495,017 – 18,495,128 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.80 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -19.49 |
| Energy contribution | -20.13 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18495017 111 + 20766785 CUAAGCAAGGCCACUUUUCAAAUGACCUCAAUUGGUU---------UUUUUUGCUGGCCUAAUGAAUAUUUCAGUUUUGGCAACAUCGCUAACGCCAUAGACGUCCAUCGAUGGCCAUUU .......((((((.....((((.((((......))))---------...)))).))))))((((........(((..((....))..)))...(((((.((......)).))))))))). ( -27.10) >DroSec_CAF1 13640 111 + 1 CUAAGCAAGGCCAAUUUUCAAAUGACCUCAAUUGGUU---------UUUUUUGCUGGCCUAAUGAAUAUUUCAGUUUUGGCAACAUCGCUAACGCCAUAGACGUCCAUCGAUGGCCAUUU .......((((((.....((((.((((......))))---------...)))).))))))((((........(((..((....))..)))...(((((.((......)).))))))))). ( -27.10) >DroSim_CAF1 2307 116 + 1 CUAAGCAAGGCCAAUUUUCAAAUGACCUCAAUUGGUUUUGUU----UUUUUUGCUGGCCUAAUGAAUAUUUCAGUUUUGGCAACAUCGCUAACGCCAUAGACGUCCAUCGAUGGCCAUUU ..((((((((((((((.((....))....)))))))))))))----).......(((((...(((.....)))((((((((............)))).))))..........)))))... ( -28.00) >DroEre_CAF1 19127 119 + 1 CUAAGCAAGUC-AACGUUUAAAUGACCUCAAUAGGUUUUCUUUUUUUUUUUUGCUGGCCUAAUGAAUAUUUCAGUUUUGGCAACAUCGCUAACGCCAUAGACGUCCAUCGAUGGCCAUAU ...((((((..-...........(((((....)))))............))))))((((...(((.....)))((((((((............)))).))))..........)))).... ( -23.71) >DroYak_CAF1 20923 93 + 1 CUAAGCAAGUC-AG------------CUCAAUAGGUUU--------UUUUUUGCUGGCCUAAUGAAUAUUUCAGUUUUGGCAACAUCG------CCAUGGACGUCUAUCGAUGGCCAUUU ...((((((..-((------------((.....)))).--------...))))))((((..............((((((((......)------))).))))(((....))))))).... ( -21.70) >consensus CUAAGCAAGGCCAAUUUUCAAAUGACCUCAAUUGGUUU________UUUUUUGCUGGCCUAAUGAAUAUUUCAGUUUUGGCAACAUCGCUAACGCCAUAGACGUCCAUCGAUGGCCAUUU ...((((((..............((((......))))............))))))((((...(((.....)))((((((((............)))).))))..........)))).... (-19.49 = -20.13 + 0.64)

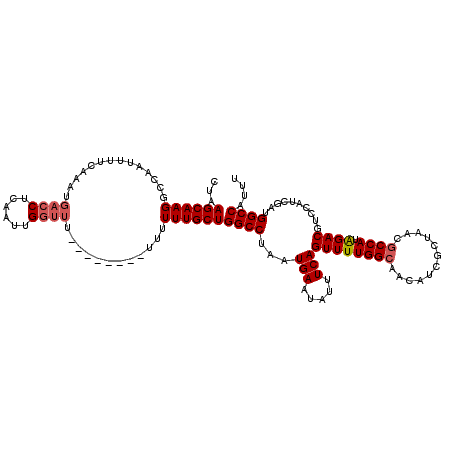
| Location | 18,495,017 – 18,495,128 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.80 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -20.01 |
| Energy contribution | -21.86 |
| Covariance contribution | 1.85 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18495017 111 - 20766785 AAAUGGCCAUCGAUGGACGUCUAUGGCGUUAGCGAUGUUGCCAAAACUGAAAUAUUCAUUAGGCCAGCAAAAAA---------AACCAAUUGAGGUCAUUUGAAAAGUGGCCUUGCUUAG ...(((((((((...(((((.....)))))..))))(((.....)))..............)))))........---------........(((((((((.....)))))))))...... ( -30.10) >DroSec_CAF1 13640 111 - 1 AAAUGGCCAUCGAUGGACGUCUAUGGCGUUAGCGAUGUUGCCAAAACUGAAAUAUUCAUUAGGCCAGCAAAAAA---------AACCAAUUGAGGUCAUUUGAAAAUUGGCCUUGCUUAG ...(((((((((...(((((.....)))))..)))))..)))).................(((((((...(((.---------.(((......)))..))).....)))))))....... ( -30.40) >DroSim_CAF1 2307 116 - 1 AAAUGGCCAUCGAUGGACGUCUAUGGCGUUAGCGAUGUUGCCAAAACUGAAAUAUUCAUUAGGCCAGCAAAAAA----AACAAAACCAAUUGAGGUCAUUUGAAAAUUGGCCUUGCUUAG ...(((((((((...(((((.....)))))..))))(((.....)))..............)))))((((....----..(((......))).(((((.........))))))))).... ( -28.80) >DroEre_CAF1 19127 119 - 1 AUAUGGCCAUCGAUGGACGUCUAUGGCGUUAGCGAUGUUGCCAAAACUGAAAUAUUCAUUAGGCCAGCAAAAAAAAAAAAGAAAACCUAUUGAGGUCAUUUAAACGUU-GACUUGCUUAG .....(((((.(((....))).)))))(((((((.(((((((.((..(((.....))))).)).)))))..........(((..((((....))))..)))...))))-)))........ ( -27.10) >DroYak_CAF1 20923 93 - 1 AAAUGGCCAUCGAUAGACGUCCAUGG------CGAUGUUGCCAAAACUGAAAUAUUCAUUAGGCCAGCAAAAAA--------AAACCUAUUGAG------------CU-GACUUGCUUAG ...(((((...(((....)))..(((------((....)))))..................)))))........--------........((((------------(.-.....))))). ( -20.00) >consensus AAAUGGCCAUCGAUGGACGUCUAUGGCGUUAGCGAUGUUGCCAAAACUGAAAUAUUCAUUAGGCCAGCAAAAAA________AAACCAAUUGAGGUCAUUUGAAAAUUGGCCUUGCUUAG ...(((((((((...(((((.....)))))..))))(((.....)))..............))))).........................(((((((.........)))))))...... (-20.01 = -21.86 + 1.85)

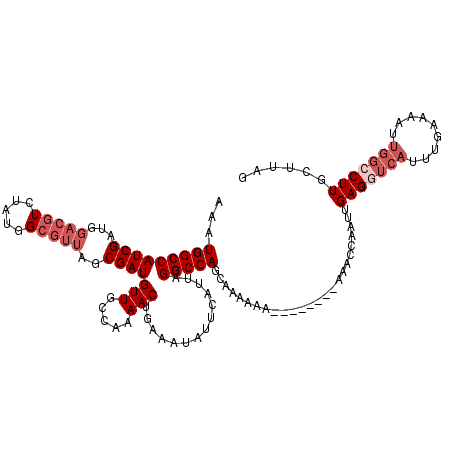

| Location | 18,495,054 – 18,495,168 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.16 |
| Mean single sequence MFE | -24.46 |
| Consensus MFE | -20.60 |
| Energy contribution | -21.04 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18495054 114 + 20766785 ------UUUUUUGCUGGCCUAAUGAAUAUUUCAGUUUUGGCAACAUCGCUAACGCCAUAGACGUCCAUCGAUGGCCAUUUCCAGCUCGAUCCAGCUGCUCCUCCAUCUCAAUCAGCCUUU ------......(((((...((((........(((..((....))..)))...(((((.((......)).))))))))).)))))........((((...............)))).... ( -24.26) >DroSec_CAF1 13677 114 + 1 ------UUUUUUGCUGGCCUAAUGAAUAUUUCAGUUUUGGCAACAUCGCUAACGCCAUAGACGUCCAUCGAUGGCCAUUUCCAGCUCGAUCCAGCUGCUCCUCCAUCUCAAUCAGCCUUU ------......(((((...((((........(((..((....))..)))...(((((.((......)).))))))))).)))))........((((...............)))).... ( -24.26) >DroSim_CAF1 2347 116 + 1 UU----UUUUUUGCUGGCCUAAUGAAUAUUUCAGUUUUGGCAACAUCGCUAACGCCAUAGACGUCCAUCGAUGGCCAUUUCCAGCUCUAUCCAGCUGCUCCUCCAUCUCAAUCAGCCUUU ..----......(((((...((((........(((..((....))..)))...(((((.((......)).))))))))).)))))........((((...............)))).... ( -24.26) >DroEre_CAF1 19166 120 + 1 UUUUUUUUUUUUGCUGGCCUAAUGAAUAUUUCAGUUUUGGCAACAUCGCUAACGCCAUAGACGUCCAUCGAUGGCCAUAUCCAGCUGGAUCCAGCUGCUCCUCCAUCUCAAUCAGCCUUU ............(((((((...(((.....))).....)))......((....))..............(((((.......((((((....)))))).....))))).....)))).... ( -28.30) >DroYak_CAF1 20948 108 + 1 ------UUUUUUGCUGGCCUAAUGAAUAUUUCAGUUUUGGCAACAUCG------CCAUGGACGUCUAUCGAUGGCCAUUUCCCACUCGAUCCAGCUGCUCCUCCAUCUCAAUCAGCCUUU ------......(((((................((((((((......)------))).))))....((((((((.......))).))))))))))......................... ( -21.20) >consensus ______UUUUUUGCUGGCCUAAUGAAUAUUUCAGUUUUGGCAACAUCGCUAACGCCAUAGACGUCCAUCGAUGGCCAUUUCCAGCUCGAUCCAGCUGCUCCUCCAUCUCAAUCAGCCUUU ............(((((................((((((((............)))).)))).......(((((.......(((((......))))).....)))))....))))).... (-20.60 = -21.04 + 0.44)

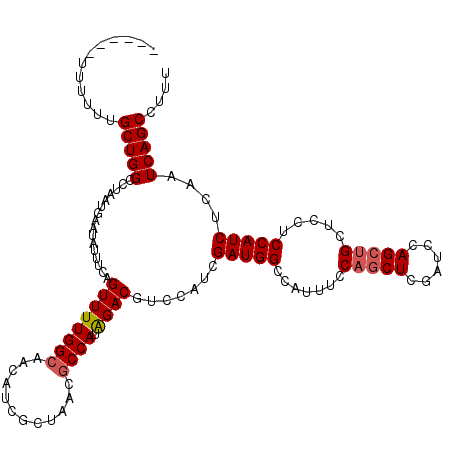

| Location | 18,495,054 – 18,495,168 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.16 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -31.58 |
| Energy contribution | -32.42 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.61 |
| SVM RNA-class probability | 0.999928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18495054 114 - 20766785 AAAGGCUGAUUGAGAUGGAGGAGCAGCUGGAUCGAGCUGGAAAUGGCCAUCGAUGGACGUCUAUGGCGUUAGCGAUGUUGCCAAAACUGAAAUAUUCAUUAGGCCAGCAAAAAA------ ...((((...((((((...((..(((((......)))))....(((((((((...(((((.....)))))..)))))..))))...))...)).))))...)))).........------ ( -35.50) >DroSec_CAF1 13677 114 - 1 AAAGGCUGAUUGAGAUGGAGGAGCAGCUGGAUCGAGCUGGAAAUGGCCAUCGAUGGACGUCUAUGGCGUUAGCGAUGUUGCCAAAACUGAAAUAUUCAUUAGGCCAGCAAAAAA------ ...((((...((((((...((..(((((......)))))....(((((((((...(((((.....)))))..)))))..))))...))...)).))))...)))).........------ ( -35.50) >DroSim_CAF1 2347 116 - 1 AAAGGCUGAUUGAGAUGGAGGAGCAGCUGGAUAGAGCUGGAAAUGGCCAUCGAUGGACGUCUAUGGCGUUAGCGAUGUUGCCAAAACUGAAAUAUUCAUUAGGCCAGCAAAAAA----AA ...((((...((((((...((..(((((......)))))....(((((((((...(((((.....)))))..)))))..))))...))...)).))))...)))).........----.. ( -35.50) >DroEre_CAF1 19166 120 - 1 AAAGGCUGAUUGAGAUGGAGGAGCAGCUGGAUCCAGCUGGAUAUGGCCAUCGAUGGACGUCUAUGGCGUUAGCGAUGUUGCCAAAACUGAAAUAUUCAUUAGGCCAGCAAAAAAAAAAAA ...((((...((((((...((..((((((....))))))....(((((((((...(((((.....)))))..)))))..))))...))...)).))))...))))............... ( -39.60) >DroYak_CAF1 20948 108 - 1 AAAGGCUGAUUGAGAUGGAGGAGCAGCUGGAUCGAGUGGGAAAUGGCCAUCGAUAGACGUCCAUGG------CGAUGUUGCCAAAACUGAAAUAUUCAUUAGGCCAGCAAAAAA------ ...((((((((.((.((......)).)).)))).(((.((..((.(((((.(((....))).))))------).))....))...))).............)))).........------ ( -27.50) >consensus AAAGGCUGAUUGAGAUGGAGGAGCAGCUGGAUCGAGCUGGAAAUGGCCAUCGAUGGACGUCUAUGGCGUUAGCGAUGUUGCCAAAACUGAAAUAUUCAUUAGGCCAGCAAAAAA______ ...((((...((((((...((..(((((......)))))....(((((((((...(((((.....)))))..)))))..))))...))...)).))))...))))............... (-31.58 = -32.42 + 0.84)

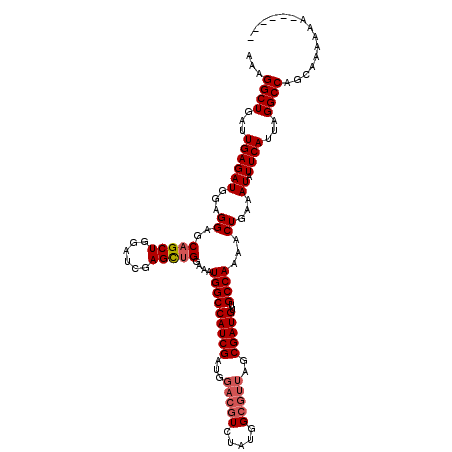

| Location | 18,495,088 – 18,495,208 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -23.15 |
| Consensus MFE | -18.62 |
| Energy contribution | -19.26 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18495088 120 + 20766785 CAACAUCGCUAACGCCAUAGACGUCCAUCGAUGGCCAUUUCCAGCUCGAUCCAGCUGCUCCUCCAUCUCAAUCAGCCUUUGGCUCAUACAUAACCCAGUAAAUCCUUUUCCAGCUCGAUG ...(((((((...((((.((.(.......(((((.......(((((......))))).....))))).......).)).))))...(((........)))...........)))..)))) ( -23.04) >DroSec_CAF1 13711 120 + 1 CAACAUCGCUAACGCCAUAGACGUCCAUCGAUGGCCAUUUCCAGCUCGAUCCAGCUGCUCCUCCAUCUCAAUCAGCCUUUGGCUCAUACAUAACCCAGUAAAUCCUUUUCCAGCUCGAUG ...(((((((...((((.((.(.......(((((.......(((((......))))).....))))).......).)).))))...(((........)))...........)))..)))) ( -23.04) >DroSim_CAF1 2383 120 + 1 CAACAUCGCUAACGCCAUAGACGUCCAUCGAUGGCCAUUUCCAGCUCUAUCCAGCUGCUCCUCCAUCUCAAUCAGCCUUUGGCUCAUACAUAACCCAGUAAAUCCUUUUCCAGCUCGAUG ...(((((((...((((.((.(.......(((((.......(((((......))))).....))))).......).)).))))...(((........)))...........)))..)))) ( -23.04) >DroEre_CAF1 19206 120 + 1 CAACAUCGCUAACGCCAUAGACGUCCAUCGAUGGCCAUAUCCAGCUGGAUCCAGCUGCUCCUCCAUCUCAAUCAGCCUUUGGCUCAUACAUAACCCAGUAAAUCCUUUUCCAGCUCGAUG ...(((((((...((((.((.(.......(((((.......((((((....)))))).....))))).......).)).))))...(((........)))...........)))..)))) ( -27.74) >DroYak_CAF1 20982 114 + 1 CAACAUCG------CCAUGGACGUCUAUCGAUGGCCAUUUCCCACUCGAUCCAGCUGCUCCUCCAUCUCAAUCAGCCUUUGGCUCAUACAUAACCCGGUAAAUCCUUUUCCAGCUCUAUG ........------.((((((.....((((((((.......))).)))))...((((................((((...))))............((.....)).....)))))))))) ( -18.90) >consensus CAACAUCGCUAACGCCAUAGACGUCCAUCGAUGGCCAUUUCCAGCUCGAUCCAGCUGCUCCUCCAUCUCAAUCAGCCUUUGGCUCAUACAUAACCCAGUAAAUCCUUUUCCAGCUCGAUG ...(((((.....((((.((.(.......(((((.......(((((......))))).....))))).......).)).))))...(((........)))...............))))) (-18.62 = -19.26 + 0.64)

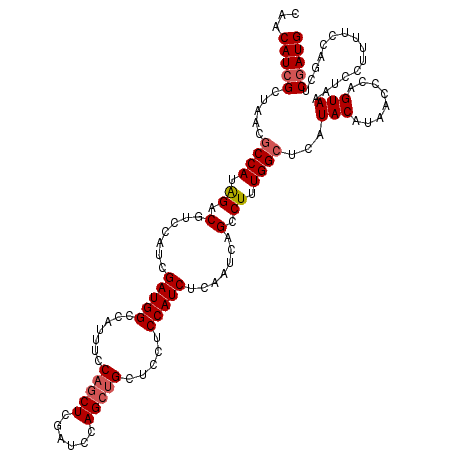
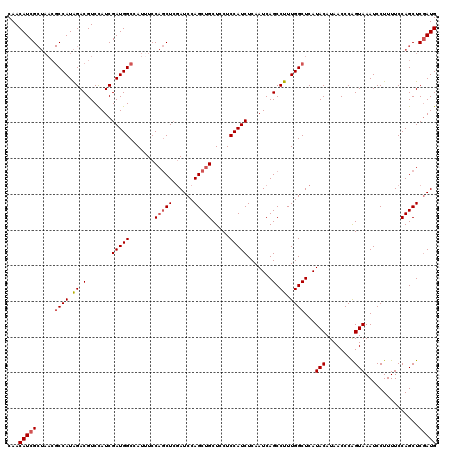
| Location | 18,495,128 – 18,495,248 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -21.62 |
| Consensus MFE | -18.40 |
| Energy contribution | -18.80 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18495128 120 + 20766785 CCAGCUCGAUCCAGCUGCUCCUCCAUCUCAAUCAGCCUUUGGCUCAUACAUAACCCAGUAAAUCCUUUUCCAGCUCGAUGGCAUCGUUGCCAAUUUUCACACCAGCGACAAUUACUCAAA ...(((.((((.(((((................((((...))))..(((........)))..........))))).)))((((....))))..........).))).............. ( -21.10) >DroSec_CAF1 13751 120 + 1 CCAGCUCGAUCCAGCUGCUCCUCCAUCUCAAUCAGCCUUUGGCUCAUACAUAACCCAGUAAAUCCUUUUCCAGCUCGAUGGCAUCGUUGCCAAUUUUCACACCAGCGACAAUUACUCAAA ...(((.((((.(((((................((((...))))..(((........)))..........))))).)))((((....))))..........).))).............. ( -21.10) >DroSim_CAF1 2423 120 + 1 CCAGCUCUAUCCAGCUGCUCCUCCAUCUCAAUCAGCCUUUGGCUCAUACAUAACCCAGUAAAUCCUUUUCCAGCUCGAUGGCAUCGUUGCCAAUUUUCACACCAGCGACAAUUACUCAAA ...(((..(((.(((((................((((...))))..(((........)))..........))))).)))((((....))))............))).............. ( -20.30) >DroEre_CAF1 19246 120 + 1 CCAGCUGGAUCCAGCUGCUCCUCCAUCUCAAUCAGCCUUUGGCUCAUACAUAACCCAGUAAAUCCUUUUCCAGCUCGAUGGCAUCGUUGCCAAUUUUCACACCAGCGACAAUUACUCAAA ...((((((((.(((((................((((...))))..(((........)))..........))))).)))((((....))))..........))))).............. ( -27.60) >DroYak_CAF1 21016 120 + 1 CCCACUCGAUCCAGCUGCUCCUCCAUCUCAAUCAGCCUUUGGCUCAUACAUAACCCGGUAAAUCCUUUUCCAGCUCUAUGGCAUCGUUGCCAAUUUUCACACCAGCGACAAUUACUCAAA .....(((....(((((................((((...))))............((.....)).....)))))...(((((....))))).............)))............ ( -18.00) >consensus CCAGCUCGAUCCAGCUGCUCCUCCAUCUCAAUCAGCCUUUGGCUCAUACAUAACCCAGUAAAUCCUUUUCCAGCUCGAUGGCAUCGUUGCCAAUUUUCACACCAGCGACAAUUACUCAAA ...(((..(((.(((((................((((...))))..(((........)))..........))))).)))((((....))))............))).............. (-18.40 = -18.80 + 0.40)

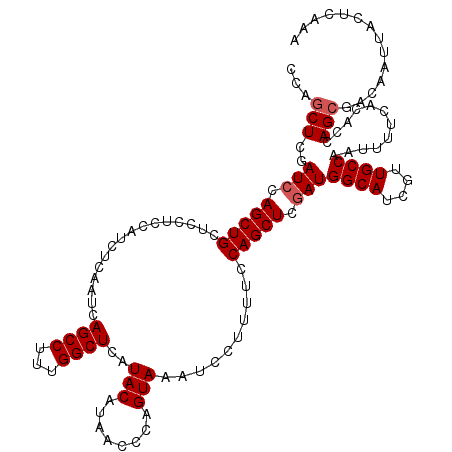
| Location | 18,495,248 – 18,495,355 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.59 |
| Mean single sequence MFE | -35.02 |
| Consensus MFE | -22.38 |
| Energy contribution | -24.75 |
| Covariance contribution | 2.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18495248 107 - 20766785 GAGCUGUGACUGCGGGGGCGGG-------CGGGAAAAGUCCGGGAAAAGCGGGUCAGGGAAAAGGGAGCGAGGACGAGGACAGUGGUAACCCAUGUUUUUGUUGUUGCCAACGG ...((.(((((((.....((((-------(.......)))))......)).))))).))......(.((((.((((((((((.(((....))))))))))))).)))))..... ( -36.30) >DroSec_CAF1 13871 114 - 1 GAGCUGUGACUGCGGGGGCGGGAAAAGUCCGGGAAAAGUCCGGGAAAAGCGGGUCAGAGAGAAGGAAGCGAGGACGAGGACAGUGGUGACCCAUGUUCUUGUUGUUGCCAACGG ((.((((..((((....))))......(((.(((....))).)))...)))).))............((((.((((((((((.(((....))))))))))))).))))...... ( -40.70) >DroSim_CAF1 2543 92 - 1 GAGCUGUGACUGCGGGGGCGGGA----------------------AAAGCGGGUCAGGGAGAAGGGAGCGAGGACGAGGACAGUGGUGACCCAUGUUUUUGUUGUUGCCAACGG ((.((((..((((....))))..----------------------...)))).))..........(.((((.((((((((((.(((....))))))))))))).)))))..... ( -30.50) >DroEre_CAF1 19366 105 - 1 GAGCUCUGACUGCGGGGGCGGGA-UGGACUGGUAAAAGUCCUGGAAAAGCGGGUCAGGGAAAAGGGGGAG----UGAGGACAG----GAGCCAUGUUUUUGUUGUUGCCAACGG ...((((((((((.....(((((-(............)))))).....)).)))))))).....(..(.(----..(.(((((----((((...))))))))).)..))..).. ( -32.60) >consensus GAGCUGUGACUGCGGGGGCGGGA______CGGGAAAAGUCCGGGAAAAGCGGGUCAGGGAAAAGGGAGCGAGGACGAGGACAGUGGUGACCCAUGUUUUUGUUGUUGCCAACGG ((.((((..((((....)))).......((((.......)))).....)))).))........(...((((.((((((((((.(((....))))))))))))).))))...).. (-22.38 = -24.75 + 2.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:59 2006