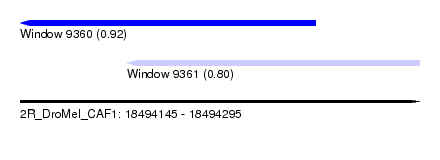
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,494,145 – 18,494,295 |
| Length | 150 |
| Max. P | 0.915552 |

| Location | 18,494,145 – 18,494,256 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.96 |
| Mean single sequence MFE | -27.84 |
| Consensus MFE | -19.94 |
| Energy contribution | -20.34 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18494145 111 - 20766785 ACAAAGCCGCUCUAAGCGAAAAGAAUCGAUGCGGCUGCUUAAAUUAUACUUUAUAAUUUGGGCAUAUA---------UGUGGAUUACCCCAAUUGGCAAUUGAACGUAUAAUAGUUCGGU ....((((((......(((......)))..))))))((((((((((((...)))))))))))).....---------(((.((((.....)))).)))(((((((........))))))) ( -31.10) >DroSec_CAF1 12854 108 - 1 GCAAAGCCGCUCUAAGCGAAAAGAAUCGAUUCGGCUGCUUAAAUUAUACUUUAUAAUUUGGGCAUAUA---------UGUGGAUUACCU---AUGGCAAUUGAACGUAUAAUAGUUCGGU .....(((((.....))......((((.((.....(((((((((((((...)))))))))))))....---------.)).))))....---..))).(((((((........))))))) ( -27.00) >DroSim_CAF1 1429 94 - 1 GCAAAGCCGCUCUAAGCGAAAAGAAUCGAUGCGGCUGCUUAAAUUAUACUUUAUAAUUUGGGCAUAUA---------UGUGGAUUACC---------AAUUGAACG--------UUUGGU (((.((((((......(((......)))..))))))((((((((((((...)))))))))))).....---------))).....(((---------((.......--------.))))) ( -26.80) >DroEre_CAF1 18241 110 - 1 GUAAAGCCGCUCUAAGCGAAAAGAAUCGAUGCGGCUGCUUAAAUUAUACUUUAUAAUUUGGGCAUAUAC---UCGUCUGUGGAUUACCA---ACUGCAAUUGGGAAUGUAUUAGUU---- ....((((((......(((......)))..))))))((((((((((((...))))))))))))((((.(---(((..((..(.......---.)..))..)))).)))).......---- ( -30.70) >DroYak_CAF1 20006 113 - 1 GUAAAGCCGCUCUAAGCGAAAAGAAUCGAUGCGGCUGCUUAAAUUAUACUUUAUAAUUUGGCCAUAUAUGUAUGUUAUGUGGAUUACCA---UUUGCAAUUGAAAAUAUAAUAGUU---- ....((((((......(((......)))..))))))((((((((((((...))))))))))(((((((.......))))))).......---...))...................---- ( -23.60) >consensus GCAAAGCCGCUCUAAGCGAAAAGAAUCGAUGCGGCUGCUUAAAUUAUACUUUAUAAUUUGGGCAUAUA_________UGUGGAUUACCA___AUGGCAAUUGAACGUAUAAUAGUU_GGU ....((((((......(((......)))..))))))((((((((((((...))))))))))))......................................................... (-19.94 = -20.34 + 0.40)


| Location | 18,494,185 – 18,494,295 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.23 |
| Mean single sequence MFE | -31.09 |
| Consensus MFE | -25.52 |
| Energy contribution | -25.40 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18494185 110 - 20766785 UAAUGGCUGGUGAGUUUUGUCUCAAAUCGGAGGA-CGGGUACAAAGCCGCUCUAAGCGAAAAGAAUCGAUGCGGCUGCUUAAAUUAUACUUUAUAAUUUGGGCAUAUA---------UGU ....((((((((.((((((((((...((....))-.))).))))))))))......(((......)))...)))))((((((((((((...)))))))))))).....---------... ( -28.30) >DroSec_CAF1 12891 110 - 1 UAAUGGCUGGUGAGUUUUGUCUCAAAUCGGAGGA-CGGGUGCAAAGCCGCUCUAAGCGAAAAGAAUCGAUUCGGCUGCUUAAAUUAUACUUUAUAAUUUGGGCAUAUA---------UGU ....((((((((((......))))..(((.((..-(((.(....).)))..))...)))...........))))))((((((((((((...)))))))))))).....---------... ( -29.00) >DroSim_CAF1 1452 110 - 1 UAAUGGCUGGUGAGUUUUGUCUCAAAUCGGAGGA-CGGGUGCAAAGCCGCUCUAAGCGAAAAGAAUCGAUGCGGCUGCUUAAAUUAUACUUUAUAAUUUGGGCAUAUA---------UGU ....(((((.((((......))))..(((.((..-(((.(....).)))..))...)))............)))))((((((((((((...)))))))))))).....---------... ( -28.70) >DroEre_CAF1 18274 116 - 1 UAAUGGCUGGUGAAUUUUGUCUCAAAUCAGAGGA-CGGGUGUAAAGCCGCUCUAAGCGAAAAGAAUCGAUGCGGCUGCUUAAAUUAUACUUUAUAAUUUGGGCAUAUAC---UCGUCUGU ......(((((..............))))).(((-((((((((.((((((......(((......)))..))))))((((((((((((...)))))))))))).)))))---)))))).. ( -40.34) >DroYak_CAF1 20039 120 - 1 UAAUGGCUGGUGAGUUUUGUCUCAAAUCGGGGGAGUGGGUGUAAAGCCGCUCUAAGCGAAAAGAAUCGAUGCGGCUGCUUAAAUUAUACUUUAUAAUUUGGCCAUAUAUGUAUGUUAUGU ..(((((((((..((.(((..((...(((..(((((((.(....).)))))))...)))...))..))).)).)))....((((((((...))))))))))))))............... ( -29.10) >consensus UAAUGGCUGGUGAGUUUUGUCUCAAAUCGGAGGA_CGGGUGCAAAGCCGCUCUAAGCGAAAAGAAUCGAUGCGGCUGCUUAAAUUAUACUUUAUAAUUUGGGCAUAUA_________UGU ....(((((.(((.(((((((((......)))((.(((.(....).))).))...))))))....)))...)))))((((((((((((...))))))))))))................. (-25.52 = -25.40 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:52 2006