
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,493,591 – 18,493,831 |
| Length | 240 |
| Max. P | 0.749012 |

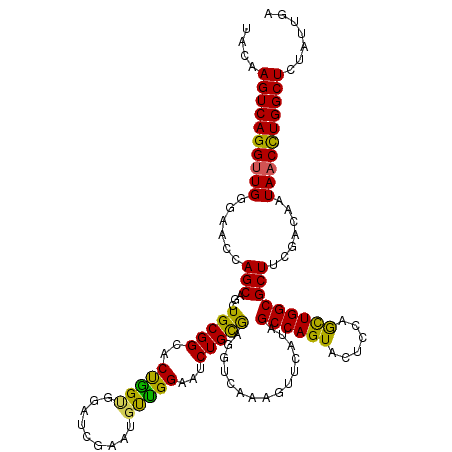
| Location | 18,493,591 – 18,493,711 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.92 |
| Mean single sequence MFE | -41.90 |
| Consensus MFE | -32.08 |
| Energy contribution | -30.68 |
| Covariance contribution | -1.40 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18493591 120 - 20766785 UACAAGUCAGGUUGGGAACCAGCAGCUGCGGCACUAGUGGAUCAAUUGCUGGAAUCUGUAAGGUCAAAGUUCAUAGCCAGUACUCCAGCUGGCGCUUUGACAAUAACUUGGCUCUCUUGA ....(((((((((((((.(((((((.(((.((....)).)..)).)))))))..))).....((((((((.....((((((......))))))))))))))..))))))))))....... ( -42.20) >DroSec_CAF1 12302 120 - 1 UACAAGUCAGGUUGGGAACCAGCAGUUGCGGCACUGGUGGAUCGAAUGUCGGAAUCUGCGAGGUCAAAGUUCAUAGCCAGUACUCCAGCUGGCGCUUCGACAAUAACCUGGCUCUAUUGA ....((((((((((.((.(((.((((......)))).))).))...(((((((....(((((.......)))...((((((......))))))))))))))).))))))))))....... ( -43.10) >DroSim_CAF1 881 120 - 1 UACAAGUCAGGUUGGGAACCAGCAGCUGCGGCACUGGUGGAUCGAAUGUCGGAAUCUGCGAGGUCAAAGUUCAUAGCCAGUACUCUAGCUGGCGCUUCGACAAUAACCUGGCUCUAUUGA ....((((((((((.((.(((.(((........))).))).))...(((((((....(((((.......)))...((((((......))))))))))))))).))))))))))....... ( -41.70) >DroEre_CAF1 17699 120 - 1 UACAAGUCAGGCUGGGAAGCAGCAGCUGCGGAACCGGCGGCUCGAAGGUUGGAAUCUGCGAGGUGAAAGUGCACAGCCAGUACUCCAGUUGGCGCUUCGACAAUAACCUGGCUCUAUUGA ....(((((((.((.(((((..((((((.((.((.(((..((((.((((....)))).))))(((......))).))).)).)).))))))..)))))..))....)))))))....... ( -42.20) >DroYak_CAF1 19471 120 - 1 UACAAGUCAGGUUGGGCACCAGCAGCUGCGGAACCGGUGGAUCGAAGUUUGGCAUCUGCGAGGUUAAAGUUCAUAGCCAGUACUCCAAUUGGCGCUUCGACAAUAACCUGGCUCUAUUGA ....((((((((((((((((.((((.(((.(((((((....)))..)))).))).))))..)))...........((((((......))))))))).......))))))))))....... ( -40.31) >consensus UACAAGUCAGGUUGGGAACCAGCAGCUGCGGCACUGGUGGAUCGAAUGUUGGAAUCUGCGAGGUCAAAGUUCAUAGCCAGUACUCCAGCUGGCGCUUCGACAAUAACCUGGCUCUAUUGA ....((((((((((......(((...(((((..(((((.........)))))...)))))...............((((((......))))))))).......))))))))))....... (-32.08 = -30.68 + -1.40)

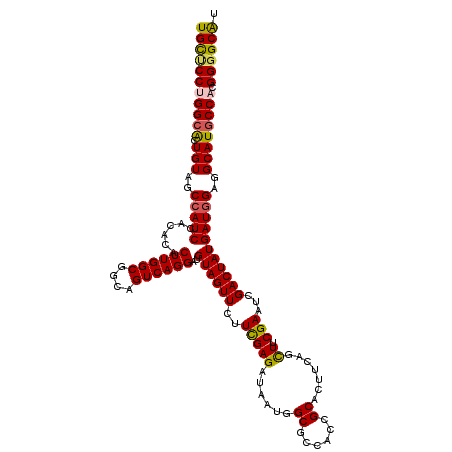
| Location | 18,493,711 – 18,493,831 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -42.68 |
| Consensus MFE | -38.90 |
| Energy contribution | -38.82 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18493711 120 + 20766785 UGCUCCUGGCGCUGUAGCCAUCCACACACUUGGCGGCAGUCAGGAUGUAGUUCUUCGAGAUAAUGGCACCACCGCACUUGAGCUUCGAAUUGACUAUGAUGGAGGCAUACCACGGUGCAU (((.(((((...(((..(((((......((((((....))))))..((((((.((((((.....((.....))((......))))))))..)))))))))))..)))..))).)).))). ( -37.90) >DroSec_CAF1 12422 120 + 1 UGCUCCUGGCACUGUAGCCAUCCACACACUUGGCGGCAGUCAGGAUGUAGUUCUUCGAGAUAAUGGCGCCACCGCACUUCAGCUUCGAAUCGACUAUGAUGGAGGCAUGCCACGGGGCAU (((((((((((.(((..(((((......((((((....))))))..((((((.((((((.....((.((....)).)).....))))))..)))))))))))..)))))))).)))))). ( -48.00) >DroSim_CAF1 1001 120 + 1 UGCUCCUGGCACUGUAGCCAUCCACACACUUGGCGGCAGUCAGGAUGUAGUUCUUCGAGAUAAUGGCGCCACCGCACUUCAGCUUCGAAUCGACUAUGAUAGAGGCAUGCCACGGGGCAU (((((((((((.(((..(.(((......((((((....))))))..((((((.((((((.....((.((....)).)).....))))))..))))))))).)..)))))))).)))))). ( -41.50) >DroEre_CAF1 17819 120 + 1 UGUUCCCGGCACUGUAGCCAUCCACACACUUGGCGGCAGUCAGGAUGUAGUUCUUCGAGAUAAUGGCGCCACCGCACUUCAGUUGCGACUGGACUAUGAUGGAGGCAUGCCACGGGGCGU .(((((.((((.(((..(((((......((((((....))))))..(((((((.(((..((...((.((....)).))...))..)))..))))))))))))..)))))))..))))).. ( -43.50) >DroYak_CAF1 19591 120 + 1 UGUCCCCGGCAUUGUAGCCAUCCACACACUUGGCGGCAGUCAGGAUGUAGUUCUUUGAGAUGAUGGCACCACCGCACUGCAGUUGCGACCUGACUAUGAUGGAGGCAUGCCAUGGGGCAU ((((((.(((((.((..(((((..((....)).....(((((((.(((((....(((((.((.(((.....))))))).)))))))).)))))))..)))))..)))))))..)))))). ( -42.50) >consensus UGCUCCUGGCACUGUAGCCAUCCACACACUUGGCGGCAGUCAGGAUGUAGUUCUUCGAGAUAAUGGCGCCACCGCACUUCAGCUUCGAAUCGACUAUGAUGGAGGCAUGCCACGGGGCAU (((((((((((.(((..(((((......((((((....))))))..((((((..(((((......((......)).......)).)))...)))))))))))..)))))))).)))))). (-38.90 = -38.82 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:50 2006