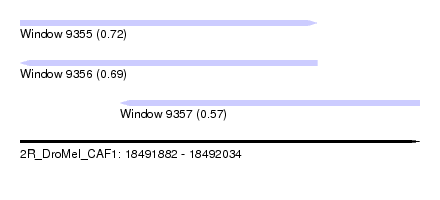
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 18,491,882 – 18,492,034 |
| Length | 152 |
| Max. P | 0.719203 |

| Location | 18,491,882 – 18,491,995 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.30 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -17.03 |
| Energy contribution | -19.09 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18491882 113 + 20766785 --AACUGAUUUUUAGAGCUGCAUGUGCGACUCUUUUUAUAGGAAAAGUUAAGUCGUUUUC-AAGGCUAGAGCUCCUUAUCUGCAAAGUCAUACCUCAAAUUGAGUGAUGGCUUUAC---- --....(((.....(((((((.((.((((((((((((....))))))...))))))...)-)..))...)))))...)))...((((((((..(((.....)))..))))))))..---- ( -29.20) >DroSec_CAF1 10489 117 + 1 --AACUGAGUUUUGGAGCUGCAUGUGCGACUCUUUUUAUAGGAAGAGUUAAGUUGUCUUC-AAGGCUAGAACUCCUUAUCUGCAAAGUCAUACCUCAAAUUGAAUGAUGGCUUUACUUCA --..((.(((((((((((.((....))(((((((((....))))))))).....).))))-))))))))..............((((((((...((.....))...))))))))...... ( -28.50) >DroEre_CAF1 15955 119 + 1 GGAACUGC-CUUUGGAGCUGCAUGUGCGACUCUUUUUAUGGGAAAUGUUAUUCAAUUUUUGGUGUGGAAUAUUUCUUAUCUGCAAAGCCAUACCUCAAAUUGAGUGAUAGCUUUGCUUCA ((.....)-)..((((((.((........(((.......)))...(((((((((((((..(((((((....((((......).))).)))))))..)))))))))))))))...)))))) ( -33.90) >DroYak_CAF1 17765 116 + 1 --AACUGC-UUUUGCAGCGGCAAGUGCAACUCUUUUUAUAGGAAAUGUUACUCCGCUUUC-CUAACUAGCACUUCUUAUCUGCAAAGUCAUAGCUCACAUUGAGUGAUGGCUUUACUUCA --......-....((((.((.((((((((......)).(((((((.((......))))))-)))....))))))))...))))((((((((.((((.....)))).))))))))...... ( -30.90) >consensus __AACUGA_UUUUGGAGCUGCAUGUGCGACUCUUUUUAUAGGAAAAGUUAAGCCGUUUUC_AAGGCUAGAACUCCUUAUCUGCAAAGUCAUACCUCAAAUUGAGUGAUGGCUUUACUUCA .............((((..((....))..))))....(((((((((((((................)))))))))))))....((((((((..(((.....)))..))))))))...... (-17.03 = -19.09 + 2.06)

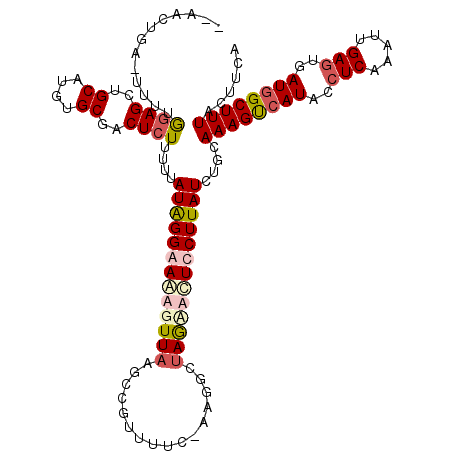
| Location | 18,491,882 – 18,491,995 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.30 |
| Mean single sequence MFE | -26.74 |
| Consensus MFE | -17.01 |
| Energy contribution | -16.70 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.689040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18491882 113 - 20766785 ----GUAAAGCCAUCACUCAAUUUGAGGUAUGACUUUGCAGAUAAGGAGCUCUAGCCUU-GAAAACGACUUAACUUUUCCUAUAAAAAGAGUCGCACAUGCAGCUCUAAAAAUCAGUU-- ----((((((.(((..(((.....)))..))).)))))).(((..((((((...((..(-(....(((((...(((((......))))))))))..)).))))))))....)))....-- ( -26.60) >DroSec_CAF1 10489 117 - 1 UGAAGUAAAGCCAUCAUUCAAUUUGAGGUAUGACUUUGCAGAUAAGGAGUUCUAGCCUU-GAAGACAACUUAACUCUUCCUAUAAAAAGAGUCGCACAUGCAGCUCCAAAACUCAGUU-- (((.((((((.(((..(((.....)))..))).))))))......((((((........-............((((((........)))))).((....)))))))).....)))...-- ( -23.60) >DroEre_CAF1 15955 119 - 1 UGAAGCAAAGCUAUCACUCAAUUUGAGGUAUGGCUUUGCAGAUAAGAAAUAUUCCACACCAAAAAUUGAAUAACAUUUCCCAUAAAAAGAGUCGCACAUGCAGCUCCAAAG-GCAGUUCC ....((((((((((..(((.....)))..))))))))))......(((.....((............(((......))).........((((.((....)).))))....)-)...))). ( -27.10) >DroYak_CAF1 17765 116 - 1 UGAAGUAAAGCCAUCACUCAAUGUGAGCUAUGACUUUGCAGAUAAGAAGUGCUAGUUAG-GAAAGCGGAGUAACAUUUCCUAUAAAAAGAGUUGCACUUGCCGCUGCAAAA-GCAGUU-- ....((((((.(((..(((.....)))..))).)))))).......((((((....(((-((((..(......).)))))))...........))))))...(((((....-))))).-- ( -29.66) >consensus UGAAGUAAAGCCAUCACUCAAUUUGAGGUAUGACUUUGCAGAUAAGAAGUUCUAGCCAU_GAAAACGAAUUAACAUUUCCUAUAAAAAGAGUCGCACAUGCAGCUCCAAAA_GCAGUU__ ....((((((.(((..(((.....)))..))).)))))).................................................((((.((....)).)))).............. (-17.01 = -16.70 + -0.31)

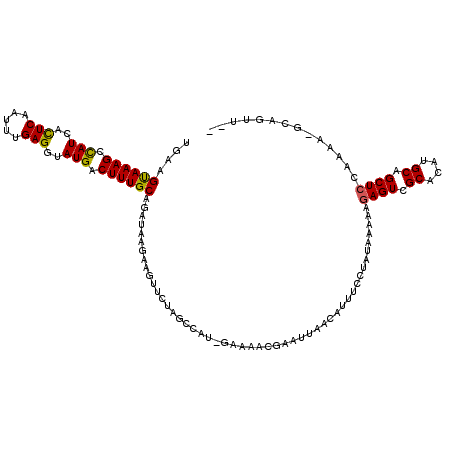
| Location | 18,491,920 – 18,492,034 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.59 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -17.69 |
| Energy contribution | -17.82 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 18491920 114 - 20766785 AGUAAUACGCAAUACUUCCUAUUUUGAUAGAGCUUCGAA-----GUAAAGCCAUCACUCAAUUUGAGGUAUGACUUUGCAGAUAAGGAGCUCUAGCCUU-GAAAACGACUUAACUUUUCC .....................(((..((((((((((...-----((((((.(((..(((.....)))..))).))))))......)))))))))...).-.)))................ ( -24.60) >DroSec_CAF1 10527 119 - 1 AGUAAUACGCAAUACUUCCUGUUUUGAUAGAGCUUCGAAGUGAAGUAAAGCCAUCAUUCAAUUUGAGGUAUGACUUUGCAGAUAAGGAGUUCUAGCCUU-GAAGACAACUUAACUCUUCC .........(((.((.....)).))).(((((((((...(..((((...(((.(((.......))))))...))))..)......))))))))).....-(((((.........))))). ( -26.00) >DroEre_CAF1 15994 120 - 1 AGUAAUAGGCAAUACUUCCUAUUUUGAUAGACCUUCGAAGUGAAGCAAAGCUAUCACUCAAUUUGAGGUAUGGCUUUGCAGAUAAGAAAUAUUCCACACCAAAAAUUGAAUAACAUUUCC .......((.((((.(((.((((((((.......))))))))..((((((((((..(((.....)))..))))))))))......))).))))))......................... ( -28.50) >DroYak_CAF1 17802 119 - 1 AGUAAUACGCAACACUUCCUACUUUGAUAGAGCUUCAAAGUGAAGUAAAGCCAUCACUCAAUGUGAGCUAUGACUUUGCAGAUAAGAAGUGCUAGUUAG-GAAAGCGGAGUAACAUUUCC .((.((.(((..((((((.((((((((.......))))))))..((((((.(((..(((.....)))..))).))))))......))))))((....))-....)))..)).))...... ( -29.00) >consensus AGUAAUACGCAAUACUUCCUAUUUUGAUAGAGCUUCGAAGUGAAGUAAAGCCAUCACUCAAUUUGAGGUAUGACUUUGCAGAUAAGAAGUUCUAGCCAU_GAAAACGAAUUAACAUUUCC ...........................(((((((((........((((((.(((..(((.....)))..))).))))))......))))))))).......................... (-17.69 = -17.82 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:55:48 2006